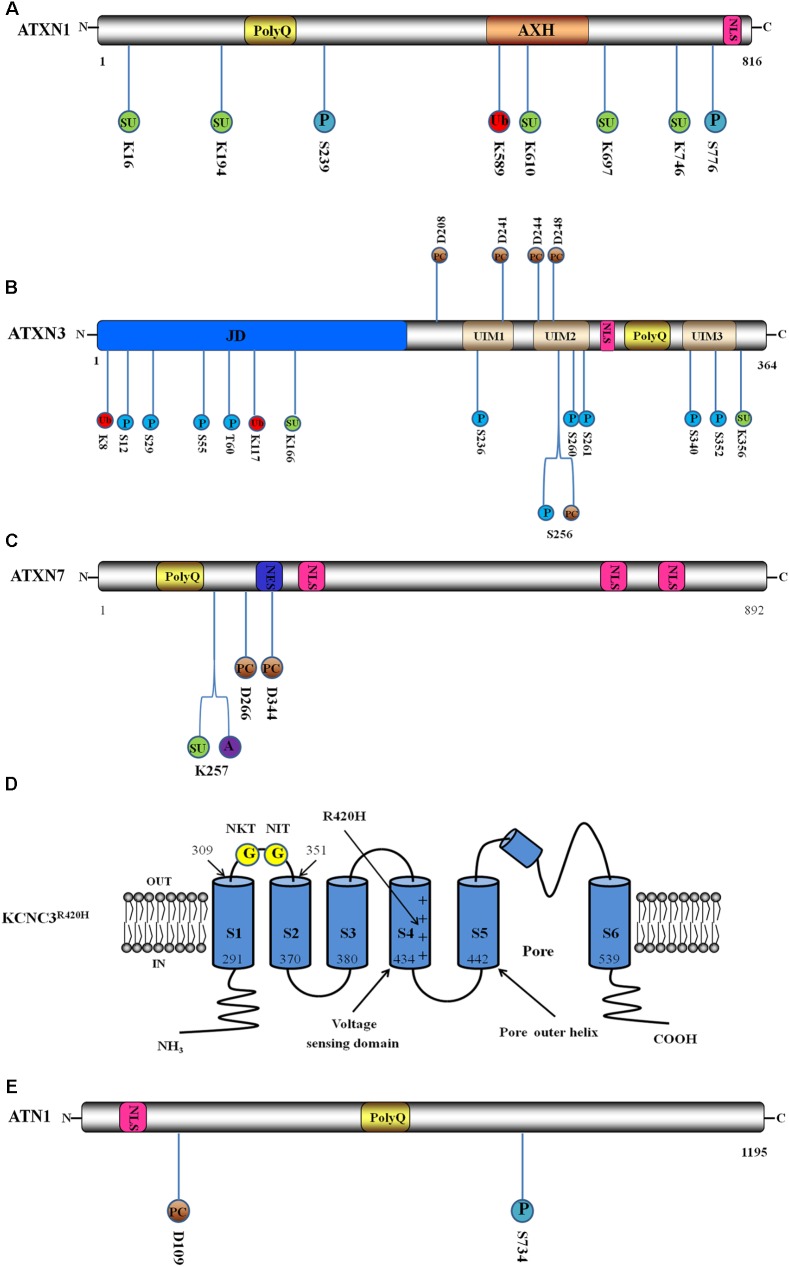
FIGURE 1.
Schematic interpretation of direct pathogenesis-related mutant protein constructs and post-translational modification (PTM) sites. Blue circle stands for phosphorylation site, red circle stands for ubiquitination site, green circle stands for SUMOylation site, brown circle represents proteolytic cleavage site, purple circle represents acetylation site, and yellow circle represents N-glycosylation site. The number below or above the circle represents the position of PTM sites. (A) Domain architecture and PTM sites of ATXN1: ATXN1 incorporates the expanded polyQ stretch, the globular ataxin1/HBP1 domain (AXH) and a nuclear localisation sequence (NLS). Here are illustrated two phosphorylation sites, a ubiquitination site and five SUMOylation sites. (B) Domain structure and PTM sites of ATXN3: ATXN3 incorporates the globular Josephin domain (JD), a flexible C-terminal tail containing three ubiquitin interaction motifs (UIMs), an NLS and the expanded polyQ. Here are illustrated ten phosphorylation sites, two ubiquitination sites, two SUMOylation sites and five proteolytic cleavage sites. (C) Domain structure and PTM sites of ATXN7: ATXN7 incorporates the expanded polyQ, a nuclear export signal (NES) and three NLS. Here are shown a SUMOylation site, an acetylation site and two proteolytic cleavage sites. (D) Schematic interpretation of the Kv3.3 channel in R420H variant (KCNC3R420H) and PTM sites. Here are illustrated the six transmembrane segments and pore re-entrant loop. The numbers show the positions of the ending and beginning amino acids of S1 and S2 domains. Location of R420H-mutation is indicated by the long arrow. The two N-glycosylation sites are located in the S1–S2 extracytoplasmic loop. Adapted from Waters et al. (2006). (E) Domain structure and PTM sites of ATN1. ATN1 incorporates an NLS and the expanded polyQ. Here are shown a phosphorylation site and a proteolytic cleavage site.