Figure 1.
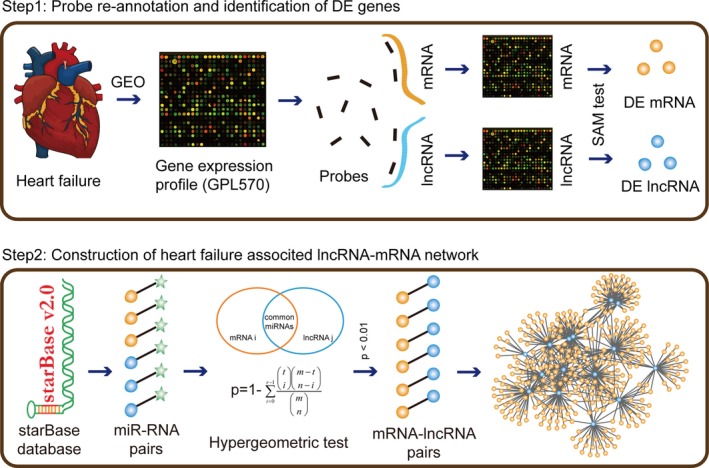
The pipeline for construction of heart failure‐related lncRNA‐mRNA network. First, we calculated the differentially expressed mRNAs and lncRNAs in normal and heart failure samples based on the re‐annotation results, using SAM test. We considered that lncRNA or mRNA with |fold change| >2 or P‐value < .01 as differentially expressed. Second, we mapped all these differentially expressed lncRNAs and mRNAs to the Ago CLIP‐supported miRNA‐target interactions and extracted the miRNA‐differentially expressed mRNAs interactions and miRNA‐differentially expressed lncRNA interactions. Third, we used hypergeometric test to identify lncRNA‐mRNA pairs at the threshold for P‐value < .01. Finally, all lncRNA‐mRNA pairs were merged to the heart failure‐related lncRNA‐mRNA network
