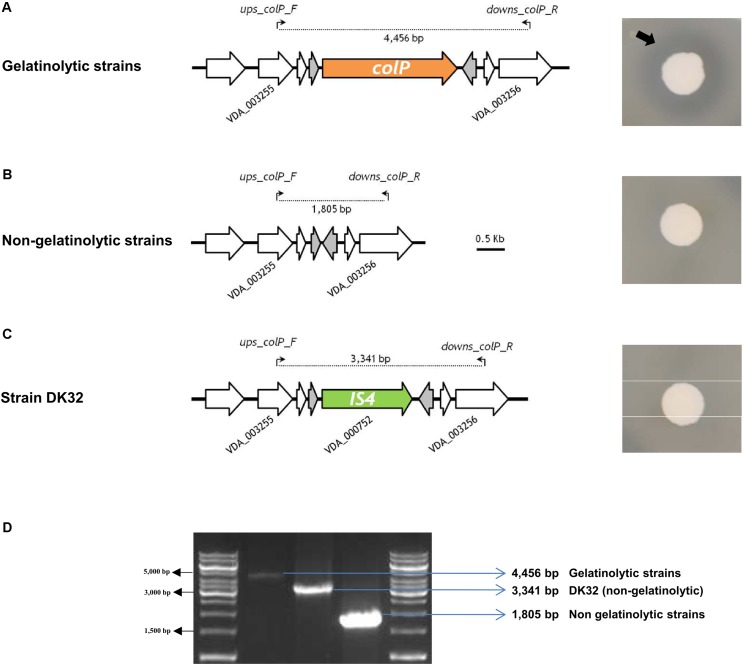
FIGURE 4.
Gelatine-degrading activity of P. damselae subsp. damselae correlates with presence of colP gene encoding a collagenase within a variable DNA region. Colony pictures at the right side of the panel depict gelatinase activity detection on agar plates. A black arrow points at the border of the translucent gelatine degradation halo. (A) Strains with capacity to degrade gelatine all test positive for internal primers for colP gene (data not shown), which is located in the same conserved genome position in all the isolates. A PCR using the primer pair ups-colp-F and downs-colp-R, designed within the flanking genes VDA_003255 and VDA_003256, respectively, produces an amplicon of 4,456 bp. (B) Gelatinase-negative strains test negative for internal primers of colP gene (data not shown), and yield a smaller amplification fragment of 1,805 bp when tested with ups-colp-F and downs-colp-R primer pair. (C) The colP-negative strain DK32 yields a different amplicon band due to the insertion of a transposase gene between VDA_003255 and VDA_003256. (D) Agarose gel electrophoresis of the three different amplicon sizes produced with primer pair ups-colp-F and downs-colp-R, revealing three different genotypes in the variable region encoding ColP collagenase.