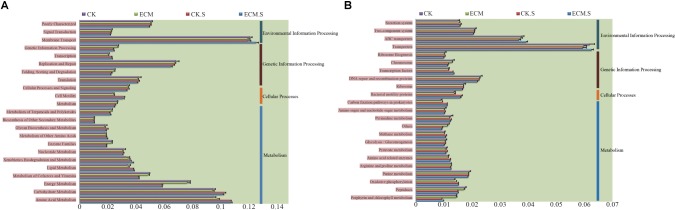
FIGURE 5.
The relative abundance of the putative metabolic functions of the bacterial community in Q. aliena roots and the surrounding soil, predicted using PICRUSt software based on the KEGG database at the second (A) and third (B) classification level. ECM, ectomycorrhizae; ECM.S, ectomycorrhizosphere soil; CK, roots of Q. aliena without T. indicum partner; CK.S, rhizosphere soil of Q. aliena without T. indicum partner. The X axis indicates the relative abundance of the metabolic functions of the prokaryotic community. Data of the above metabolic function analysis are presented as mean ± SD (error bars), calculated from the four biological replicates of each treatment group.