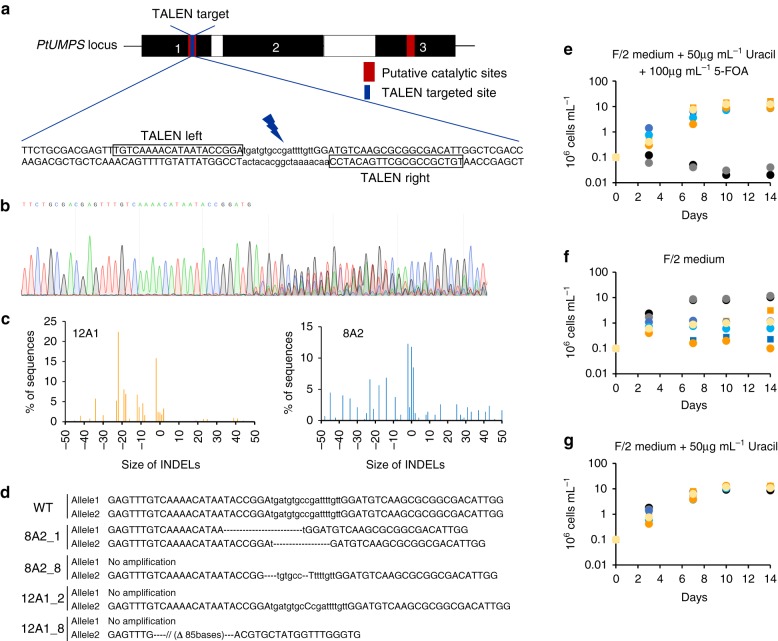
Fig. 1.
Characterization of PtUMPS knock-out strains generated using a TALE nuclease. a Structure of the PtUMPS locus. The exons are represented as black boxes, with the putative catalytic sites in red and the TALEN-targeted site in blue. The half-sites recognized by each TALEN are boxed. b Sequence chromatogram of a PCR product amplified from strain 12A1, showing the presence of heterogeneous sequences surrounding the TALEN cutting site. c INDEL spectrum and frequency predicted by the Track INDELs by DEcomposition (TIDE) algorithm in the two 5-FOA-resistant colonies, 8A2 and 12A1. d Example of mutagenic events through the alignment of the wild type and 8A2_1, 8A2_8, 12A1_2, and 12A1_8 subclones DNA sequences at the PtUMPS locus. e–g Representative growth experiments from two independent repeats performed over 14 days with the parental NCMA strain (black circles), the 2A3 control strain transformed with the NAT vector only (gray circles), two 5-FOA-resistant populations (8A2 dark blue squares and 12A1 dark orange squares), and four subclones: 8A2_1 (dark blue circles), 8A2_8 (light blue circles), 12A1_2 (dark orange circles) and 12A1_8 (yellow circles), e F/2 medium supplemented with 50 µg mL−1 uracil and 100 µg mL−1 5-FOA, f classical F/2 medium, or g F/2 supplemented with 50 µg mL−1 uracil. Y-axis e–g presents cell density, expressed as million cells per mL, on a logarithmic scale