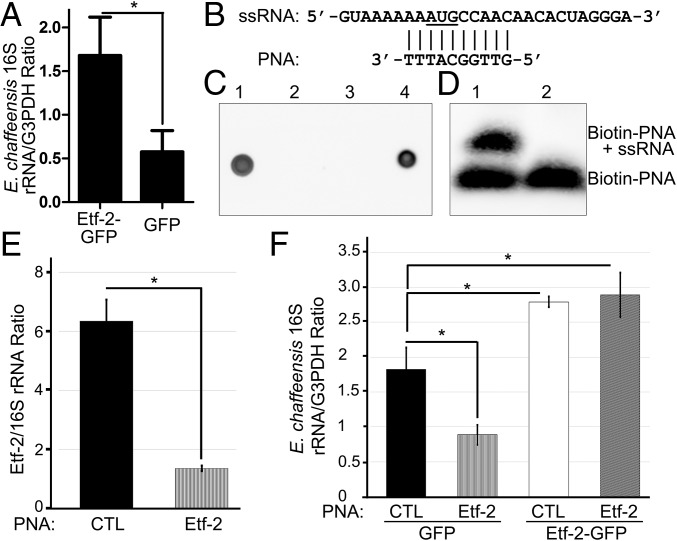
Fig. 8.
Overexpression of Etf-2 promotes ehrlichial infection, and Etf-2–specific antisense PNA inhibits ehrlichial infection. (A) HEK293 cells were transfected with Etf-1-GFP or GFP. At 8 hpt, freshly isolated E. chaffeensis was added, and cells were harvested at 2 dpi. Bacterial numbers in each sample were determined by qPCR using specific primers for Ehrlichia 16S rDNA and were normalized against the level of human G3PDH (glyceraldehyde 3-phosphate dehydrogenase). Data are presented as the mean ± SD from three independent experiments. *P < 0.05, two-tailed t test. (B–D) Etf-2 PNA binds the target ssRNA. (B) Etf-2 PNA targets the putative translation start site (underlined) of the mRNA. The upper line shows the ssRNA sequence designed for the hybridization assay. (C) Dot blot analysis of biotinylation of PNAs using HRP-conjugated streptavidin. Dots 1–4: 1, biotin-Etf-2 PNA; 2, unlabeled Etf-2 PNA; 3, unlabeled control (CTL) PNA; 4, biotin-CTL PNA. (D) EMSA of biotinylated Etf-2 PNA bound to ssRNA. Lane 1, labeled Etf-2 PNA incubated with ssRNA; lane 2, labeled Etf-2 PNA only. (E) Etf-2 PNA significantly reduces ehrlichial etf-2 mRNA expression. HEK293 cells infected with Etf-2–transfected or CTL PNA-transfected E. chaffeensis were harvested at 48 hpi and subjected to qRT-PCR analysis. The values reflect bacterial etf-2 mRNA normalized against 16S rRNA. (F) HEK293 cells expressing Etf-2 complemented the Etf-2 PNA knockdown of Ehrlichia infection. RNA samples prepared from HEK293 cells expressing Etf-2-GFP or GFP incubated with Etf-2–transfected or CTL PNA-transfected Ehrlichia were harvested at 48 hpi and subjected to qRT-PCR analysis. The values reflect bacterial 16S rRNA normalized against human G3PDH mRNA. Data indicate the mean ± SD from three independent experiments that had three replicates per sample. *P < 0.05, two-tailed t test or ANOVA.