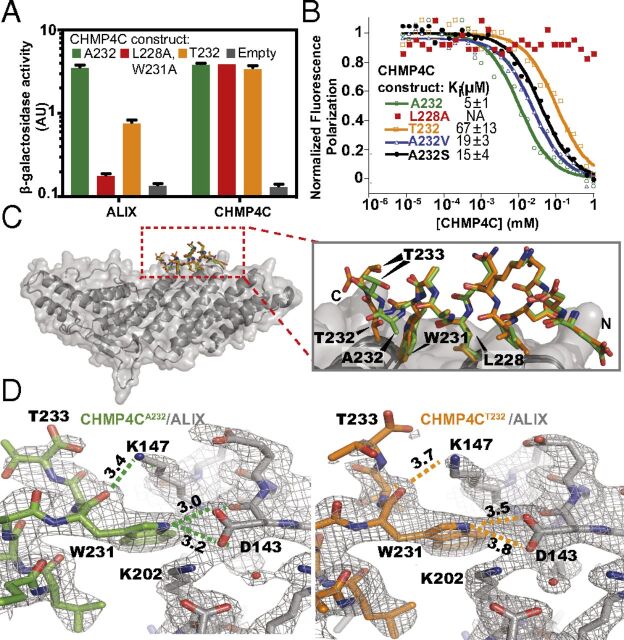
Fig. 1.
The CHMP4C A232T substitution reduces ALIX binding. (A) β-Galactosidase activity assays of yeast cotransformed with the indicated full-length CHMP4C constructs fused to VP16 and full-length ALIX or CHMP4C fused to GAL4 (mean ± SD, n = 3). (B) Competitive fluorescence polarization binding assay with an ALIX construct spanning the Bro1 and V domains (residues 1–698) binding to fluorescently labeled CHMP4CA232 peptide (residues 216–233) competing with the indicated unlabeled CHMP4C peptides. Curves are from a representative experiment. Ki values are expressed as mean ± SD. n ≥ 7. (C, Left) Superposition of ALIX Bro1 domain (gray) complexes with CHMP4CA232 (green) and CHMP4CT232 (orange) peptides. (Right) Enlarged view of the CHMP4C peptide superposition highlights the reduction in CHMP4CT232 helicity. (D) Binding interfaces between ALIX (gray) and CHMP4CA232 (green) (Left) and CHMP4CT232 (orange) (Right). Figures show equivalent intermolecular distances (in angstroms) for different atoms of CHMP4C residue W231. See also SI Appendix, Fig. S1 and Table S1.