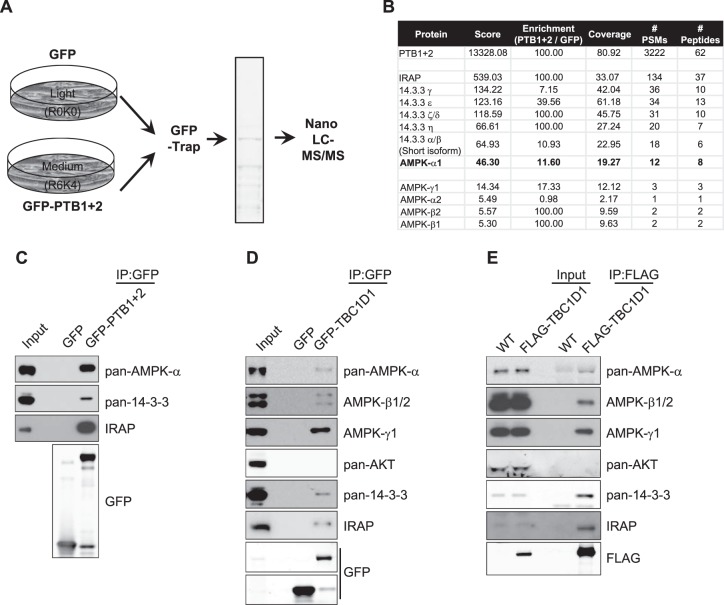
Figure 1. AMPK precipitates with TBC1D1 PTB domains in C2C12 myotubes.
(A) Schematic of the SILAC experiment set-up used to identify proteins interacting with the PTB domains of TBC1D1 in differentiated C2C12 myotubes. (B) Table of a subset of the proteomics output. Score: combines several parameters (a high score generally signifies a high protein abundance and a high confidence in the detection and quantification by the software). Enrichment: Ratio of the quantification values of the medium (GFP-PTB 1 + 2) and light (GFP) quantification channels. Coverage: percentage of the protein sequence covered by identified peptides. # PSMs (peptide spectrum matches): total number of identified peptide sequences (includes those redundantly identified). # Peptides: total number of unique identified peptides. (C) Western blot validation of proteomics results. GFP-trap precipitates from C2C12 myotube extracts, lentivirally transduced to express either GFP or GFP-PTB1 + 2 blotted for the proteins indicated. (D) Lysates from differentiated C2C12 myotubes lentivirally transduced with GFP or full-length GFP-tagged TBC1D1 (GFP-TBC1D1) subjected to GFP-trap and western blotting for the proteins indicated. (E) Homogenised quadricep lysates from either wild-type (WT) mice or mice expressing 3xFLAG-TBC1D1 were immunoprecipitated with a FLAG antibody and resulting complexes analysed. Representative blots of n = 4 independent experiments.