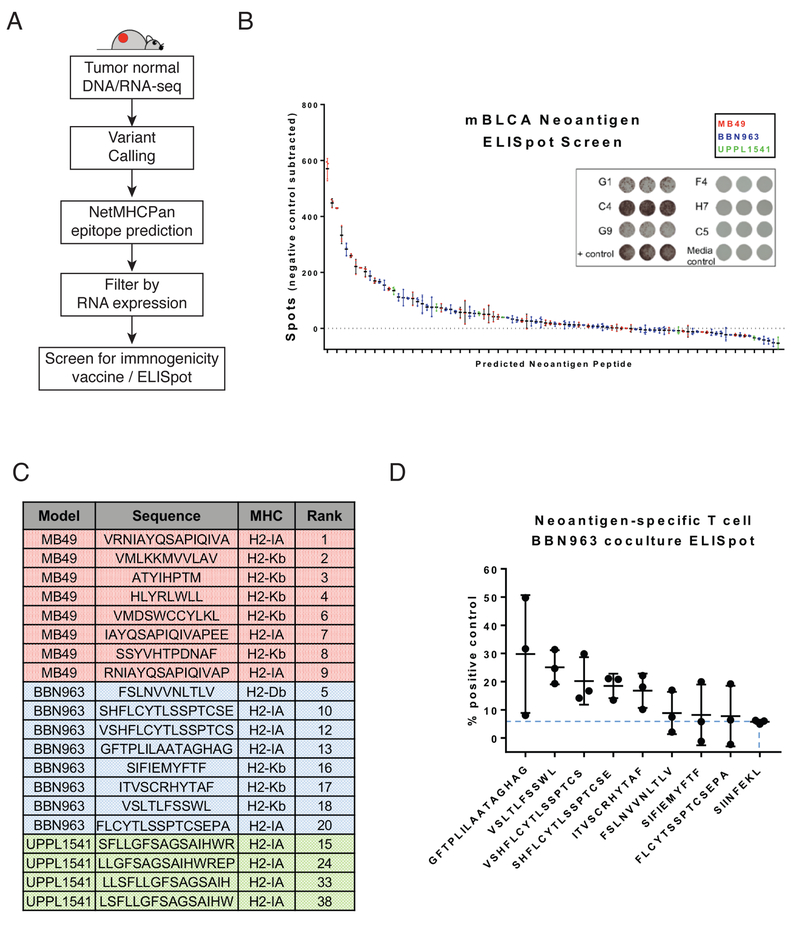
Figure 7. Neoantigen prediction and validation in BBN963, UPPL1541, and MB49.
A) Schema for neoantigen prediction workflow, using tumor DNA, tumor RNA, and matched normal DNA to call mutations via UNSeqr, epitope prediction to identify predicted class I and II binders, and vaccine/ELISpot validation. B) Summary of vaccine/ELISpot results in MB49, BBN963, and UPPL1541, with background subtracted counts ranked by number of spots and representative figures of highly immunogenic wells (G1, C4, G9), weakly immunogenic wells (F4, H7, C5), and controls. C) Summary of top eight predicted neoantigens in MB49 (red), BBN963 (blue) and top four predicted neoantigens in UPPL1541 (green), including sequence, predicted MHC class, and rank among all screened peptides within all three models. D) IFN-γ ELISpot results of BBN963 neoantigen-enriched T cells co-cultured with BBN963 tumors, as a percentage peptide-pulsed target positive control. Blue dashed line marks IFN-γ intensity of irrelevant peptide (SIINFEKL) enriched T cells co-cultured with BBN963.