FIG. 3.
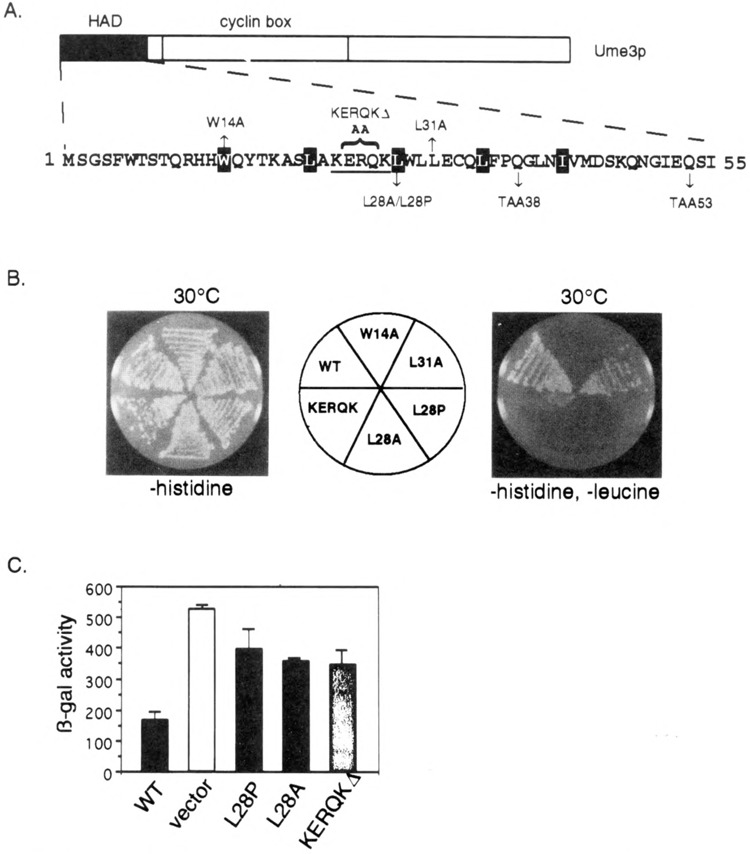
Functional analysis of the HAD mutants. (A) HAD mutant descriptions. The amino-terminal region containing the holoenzyme associating domain (HAD) is presented. Site-directed alterations are shown above and below the primary sequence. The KERQK sequence (bold) was substituted with two alanines to form the KERQKΔ mutation. The stop codons generated by the mutagenesis (TAA38 and TAA53) are shown. The reiterated hydrophobic residues comprising the putative leucine zipper are depicted as white lettering on black. (B) HAD mutations disrupt in vivo recruitment of the holoenzyme. Wild-type (WT) lexA-Ume3p or HAD mutants are tested for their ability to activate the lexAop-LEU2 reporter gene at 30°C as indicated by growth on –histidine, –leucine medium (right). No growth defect is apparent when only plasmid maintenance is selected (–histidine). Positive control is the same as Fig. 1. (C) HAD mutants are defective in Ume3p-dependent repression. The expression of ssa1-lacZ reporter plasmid was assayed in the HAD mutants and wild-type Ume3p constructs (no lexA domain) in a ume3Δ background. Three independent transformants from each group were assayed and β-galactosidase (β-gal) activity is presented in Miller units (42). Error bars indicate standard deviations of the experiments.
