Figure 1.
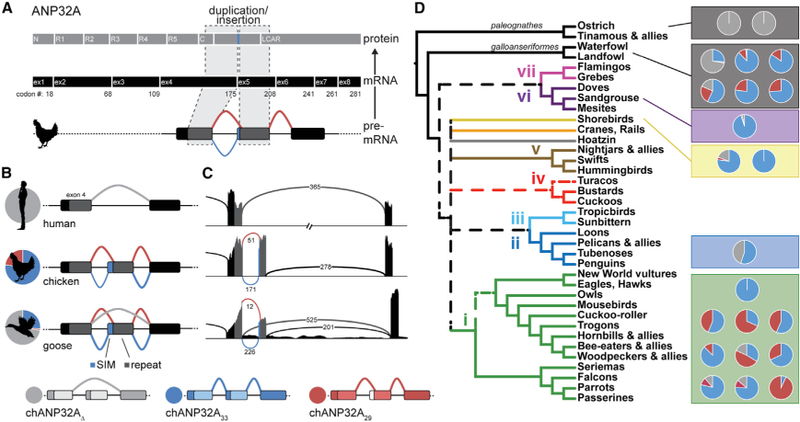
Natural Variation in ANP32A Splicing Patterns in Aves Duplication and insertion of ANP32A exon 4 results in three major splice isoforms in birds. Schematic of chANP32A protein and exonic (ex) mRNA organization indicating duplicated domains. C, C-cap; N, N-cap; R1–R5, leucine-rich regions 1–5. Exon numbering is based on chANP32A33. All human ANP32A transcripts lack the exon duplication (light gray). Schematics of chicken and goose transcripts show splicing upstream to capture codingsequence for the SIM (blue), splicing downstream to omit the SIM (red), or in some cases skipping the repeated exon to create a mammalian-like transcript (light gray). The relative abundance of each splice isoform in RNA-seq data is indicated by the pie charts. SIM, SUMO-interacting motif. Sashimi plots of ANP32A corresponding to examples in (B) and colored similarly. The abundance of each splice variant is indicated on the lines corresponding to the intron-spanning reads. ANP32A splice patterns in diverse bird species overlaid on the Aves consensus phylogeny (dashed lines represent branches that were in two of three consensus trees from Reddy et al., 2017). Pie charts represent relative transcript abundance from RNA-seq datasets (listed in Table S1).
