Figure 6.
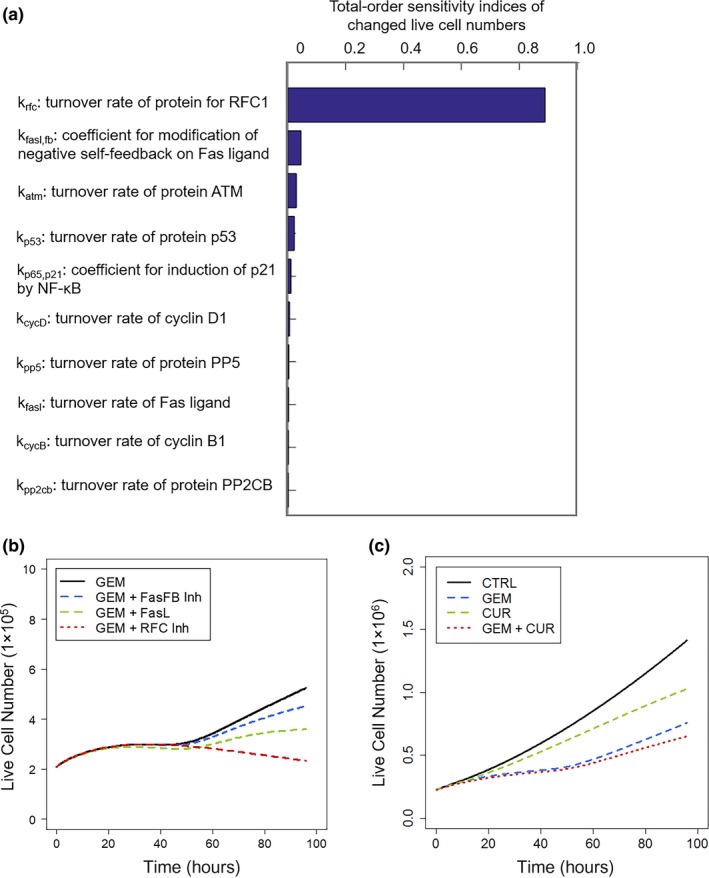
Model‐based analysis and simulations for target selection and prediction of drug combination efficacy. Sobol Sensitivity Analysis assisted in the selection of protein targets that could potentially enhance gemcitabine efficacy. (a) Rank‐order sensitivity indices of model kinetic parameters having the most significant impact on proliferating cell numbers. (b) Simulated profiles of cell proliferation responses to 20 nM gemcitabine alone (GEM; black), development of resistance to Fas pathway signaling (FasFB Inh; blue), Fas ligand exposure (FasL; green), or combined with inhibition of DNA repair (RFC Inh; red). (c) Model‐based simulation of cell proliferation profiles of drug‐free control cells (black) and when treated with gemcitabine (GEM; blue), curcumin (CUR; green), or both (red). Specific model parameters were fixed based upon assumed mechanisms of curcumin action (see text).
