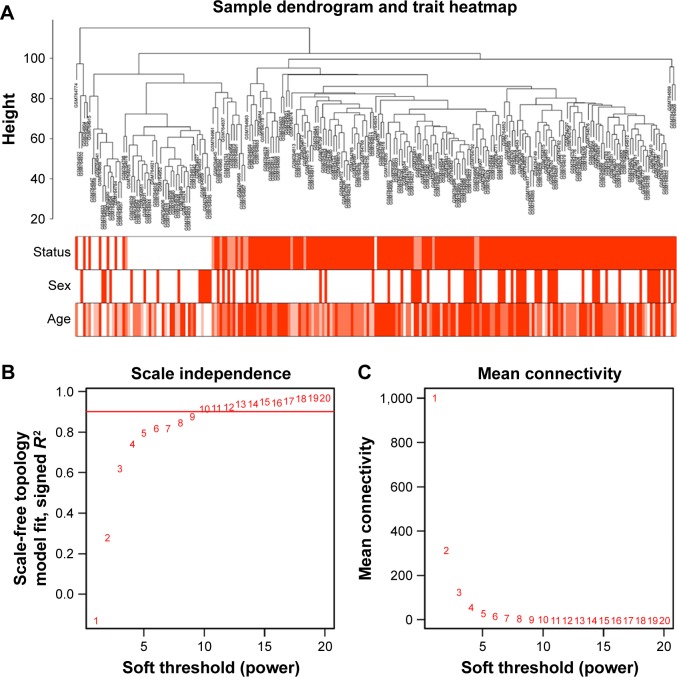
Figure 1.
Clustering of samples and determination of soft-thresholding power.
Notes: (A) The clustering was based on the expression data of GSE30784, which contained 167 OSCC, 17 dysplasia and 45 normal samples. The top 5,000 genes with the highest SD values were used for the analysis by WGCNA. The color intensity was proportional to disease status (normal, dysplasia and OSCC), sex and age. (B) Analysis of the scale-free fit index for various soft-thresholding powers (β). (C) Analysis of the mean connectivity for various soft-thresholding powers. In all, 9 was the most fit power value.
Abbreviations: OSCC, oral squamous cell carcinoma; WGCNA, weighted gene co-expression network analysis.