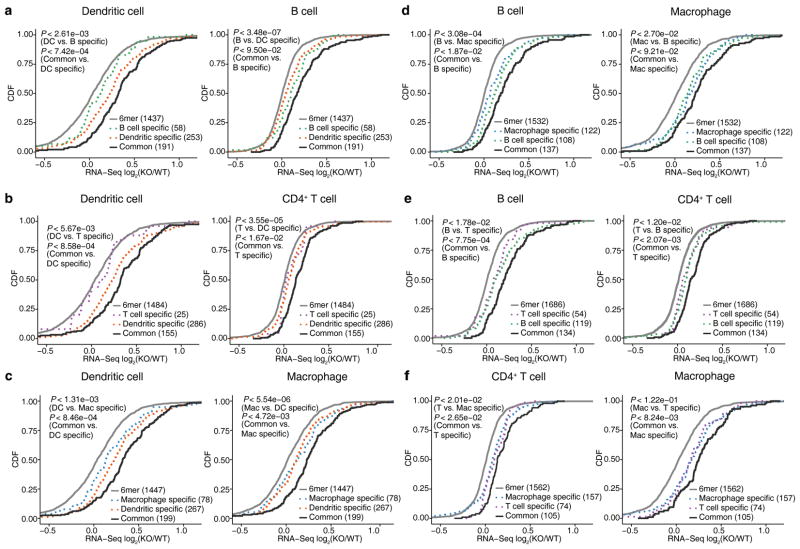
Figure 3.
Context-specific miR-155 targeting leads to differences in gene regulation between cell types. For all six pairwise comparisons across four immune cells, de-repression of genes containing common (solid lines) and cell-type specific (dotted lines) 3′UTR miR-155 dependent iCLIP sites is shown in the form of CDFs. Genes with 3′UTR miR-155 seed matches are also shown as reference. Only co-expressed genes (WT RNA-Seq FPKM > 1 and difference < 16 fold) are included in each pairwise comparison. In each plot, two P-values from one-sided KS tests are shown. First KS test corresponds to the comparison between all miR-155 target genes identified in this cell type and genes only targeted in the other cell, while the second one corresponds to the comparison between the common target genes and target genes specific to this cell type. Results of four independent iCLIP and three independent RNA-Seq experiments are shown.