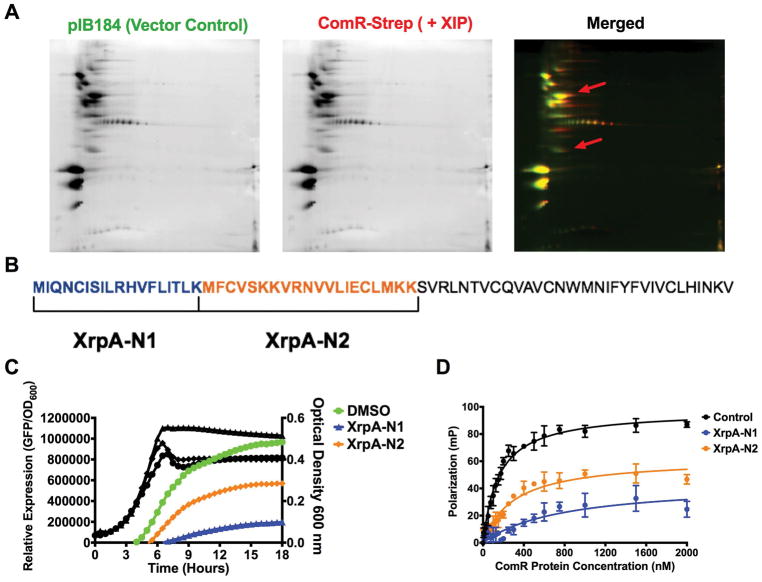
Figure 7. The N-terminus of XrpA inhibits ComR.
(A) Individual 2D gels of pIB184 (vector control; green) and ComR-Strep with 2 μM sXIP (red) elutions obtained during SPINE experiments, along with a merged image. The red arrows indicate regions where XrpA peptides were identified via spot picking and LC-MS/MS of trypsin-digested proteins. (B) Selection of XrpA-N1 and XrpA-N2 synthetic peptides based on the sequences returned by LC-MS/MS. The full length 69-aa XrpA protein is shown with each selected peptide highlighted by color and brackets. To the right is a table with the start and stop position of each peptide in XrpA, the peptide sequence, and the length in aa residues. (C) Transcriptional activation assays using a fused PcomX::gfp reporter strain in CDM medium with addition of 10 μM of either XrpA-N1 or XrpA-N2 compared to DMSO control. (D) Fluorescence polarization (FP) curves of increasing concentrations of purified ComR binding to 10 nM of PcomX dsDNA probe in the presence of 10 μM sXIP and 10 μM of either XrpA-N1 or XrpA-N2 compared to DMSO control. Kd values are shown in Table 2. Both transcriptional activation assay and FP assay results are averages from three independent experiments.