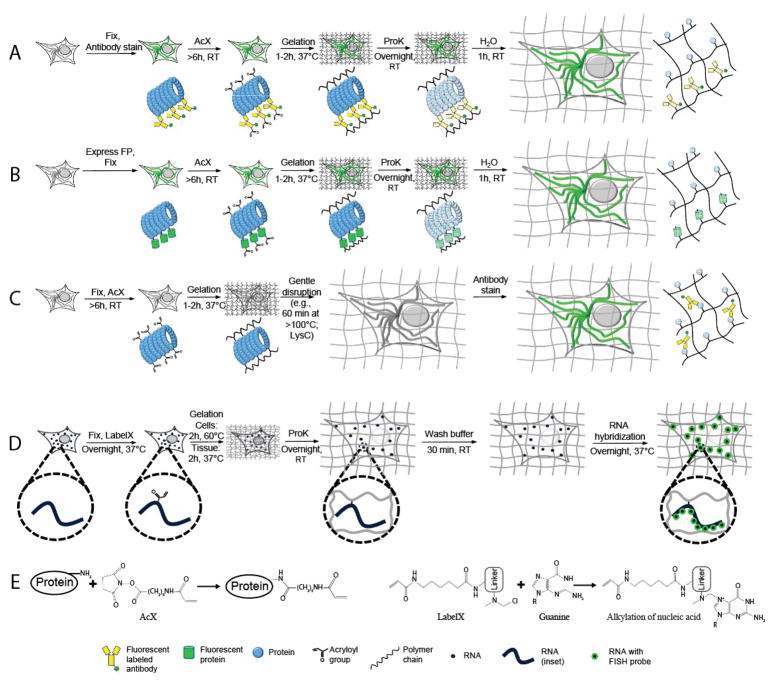
Figure 1.
Expansion microscopy (ExM) workflows discussed in this unit. (A–C) Protein retention ExM (proExM) workflows. A, Samples are fixed and stained with antibodies using conventional immunostaining protocols, then treated at room temperature (RT) with Acryloyl-X SE (AcX, see panel E, left, for detail), which enables proteins to be anchored to the hydrogel. The samples then undergo gelation, proteinase K (ProK) treatment for mechanical homogenization (also called digestion), and expansion in water. (B) Samples expressing fluorescent proteins are fixed and treated with AcX before going through gelation, mechanical homogenization, and expansion in water. (C) Samples are fixed and treated with AcX before going through gelation, a comparatively gentle mechanical homogenization process (e.g., high temperature denaturing in detergent solution), and expansion, followed by antibody staining. (D) Expansion fluorescence in situ hybridization (ExFISH). Samples are fixed and treated with LabelX (see panel E, right, for detail), which enables RNA to be anchored to the polymer. The samples then go through gelation, mechanical homogenization, and expansion. Finally, FISH probes are hybridized to the anchored RNA. (E) Schematics of AcX binding to a protein (left) and LabelX binding to a guanine base of RNA (right). Modified from (Tillberg et al., 2016) and (Chen et al., 2016).