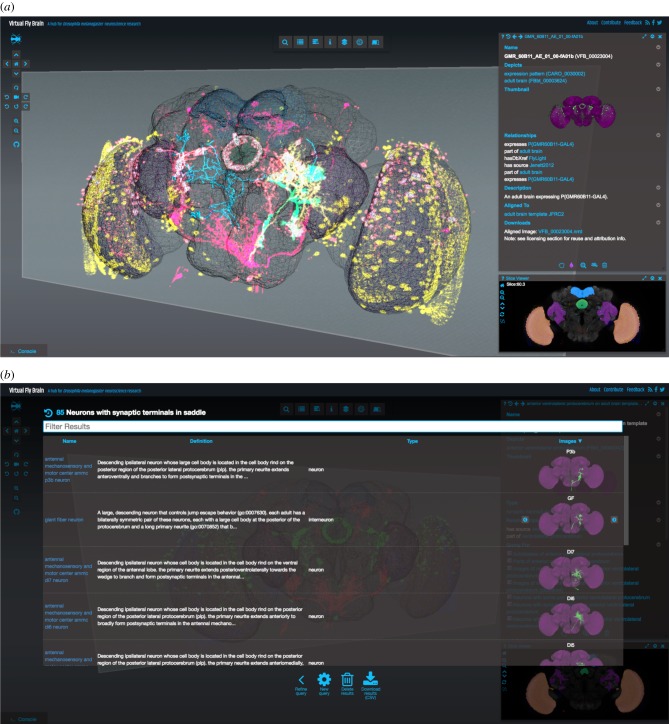
Figure 5.
(a) VFB main view shows a reference template for the D. melanogaster from Janelia Research Campus using the 3D Canvas. Superposed on the template are various gene expression patterns visualized as point clouds, reconstructed neurons and segmented neuropil regions. At the bottom right corner, the Stack Viewer shows a frontal slice through the superposed confocal microscopy images. The Stack Widget is fully synchronized with the 3D Canvas and a moving 3D plane indicates the specific slice currently displayed. On the top of the Stack Widget, a Geppetto viewer is used to display the ontological information associated with the current selection. (b) Geppetto's Query component is used to display the results of queries that can be executed from the UI, in this case, the UI shows all the neurons with synaptic terminals in the saddle. By clicking on the thumbnails, the selected neuron is loaded on demand and visualized in the 3D Canvas, the Stack Viewer and the textual definition.