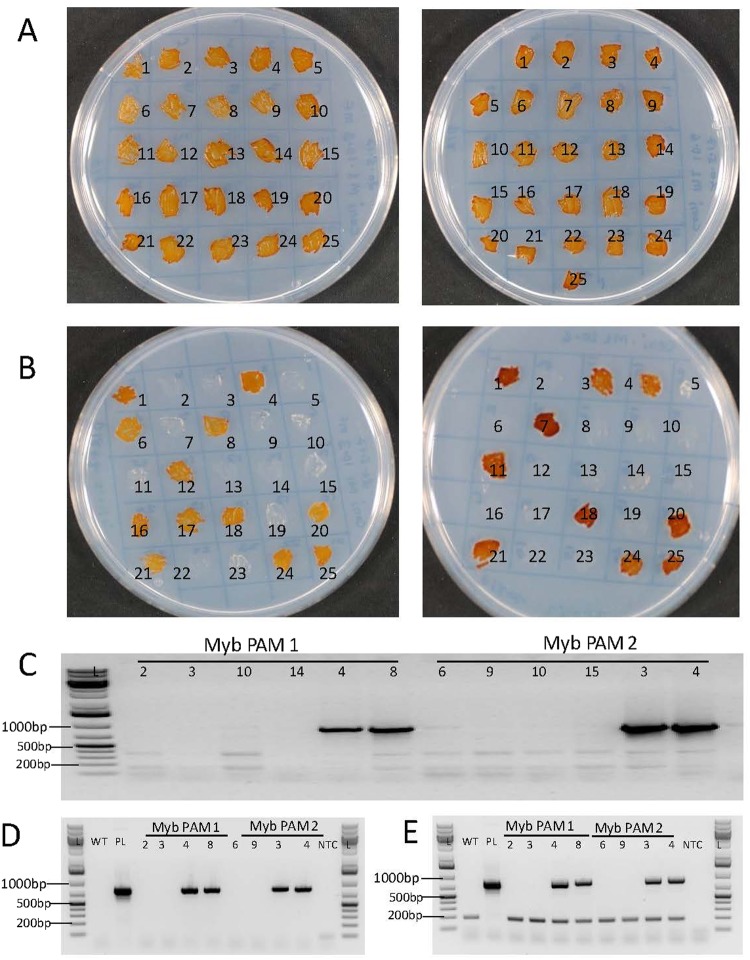
Figure 5.
Identification of non-transgenic mutants. 25 colonies from Myb PAM 1 (left side) and Myb PAM2 (right side) patched on 50% seawater (SW) f/2 agar plate without zeocin (A) and with zeocin (B). All patched colonies survived on non-selection plates whereas only colonies still containing pPtPuc3m-Cas9_sgRNA survived on selection plates. Colonies lacking the plasmid appeared white and dead after ten days on selection plates. Presented pictures were taken seven days after patching. Loss of pPtPuc3m-Cas9_sgRNA episome in colonies not able to survive on selection plates was verified by the inability to amplify a Cas9 DNA fragment (817 bp) from these colonies (C). Cas9 PCR products generated from colonies still able to grow on selection plates were included as positive controls. Ten months after initial transformation episome free colonies and colonies containing episomes were reanalyzed for presence of Cas9 DNA (817 bp fragment) (D). The origin of replication from the episome (855 bp fragment) and a genomic control fragment from Phatr2_52110 (188 bp) were co-amplified as a control of the samples (E). Lanes marked L contains GeneRuler 1 kb Plus DNA Ladder (Fermentas), WT is wild type P. tricornutum, PL is pPtPuc3m-Cas9_sgRNA plasmid and NTC (non-template control) is water. Numbers above lanes correspond to colony numbers indicated on the agar plates.