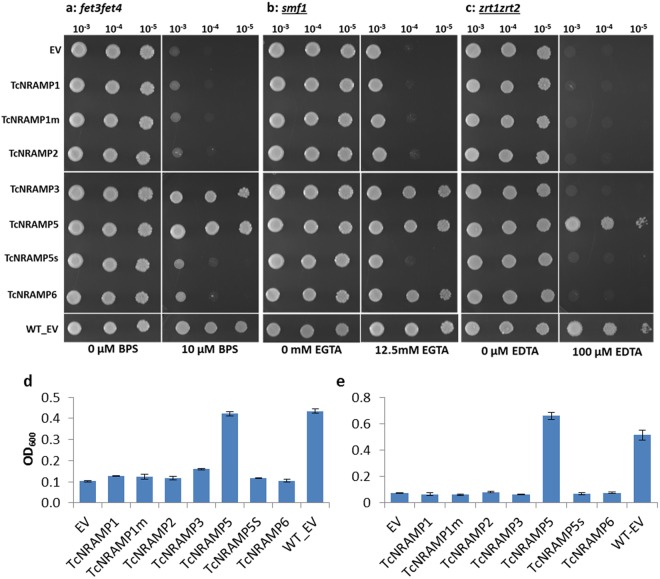
Figure 7.
Functional analysis of cacao NRAMP genes in yeast. Functional complementation for uptake of Fe2+, Mn2+ and Zn2+ in the yeast strains DEY1453 (fet3fet4), YOL122C (smf1) and ZHY3 (zrt1zrt2). Different yeast strains expressing TcNRAMP1, 1m (mutated version of TcNRAMP1), 2, 3, 5, 5s (splice variant of TcNRAMP5), 6 or empty vector pDR195 (EV) were cultured on synthetic defined (SD)-Ura plates containing 2% glucose in presence or absence of respective metal chelator. Transformed fet3fet4 cells were spotted on the plate (pH 4.0) supplemented with 10 μM FeCl3 or 10 μM Fe chelator (a) and also grown (d) in low iron liquid medium (LIM10). Transformed smf1 cells were grown on the plate (pH 5.2) supplemented with 100 μM MnSO4 or with 12.5 mM Mn chelator EGTA (b). The yeast mutant strain ZHY3 (zrt1zrt2) expressing the genes were assessed on the plate (pH 5.8) supplemented with 100 μM ZnCl2 or with 100 μM of metal chelator EDTA, 10 μM each of FeCl3 and ZnCl2 (c) and also grown (e) in low zinc liquid medium (LZM100). The wild type yeast strain DY1457 transformed with the empty vector pDR195 (WT-EV) was included as a positive control. Data are means ± SD of three biological replicates.