FIG 6.
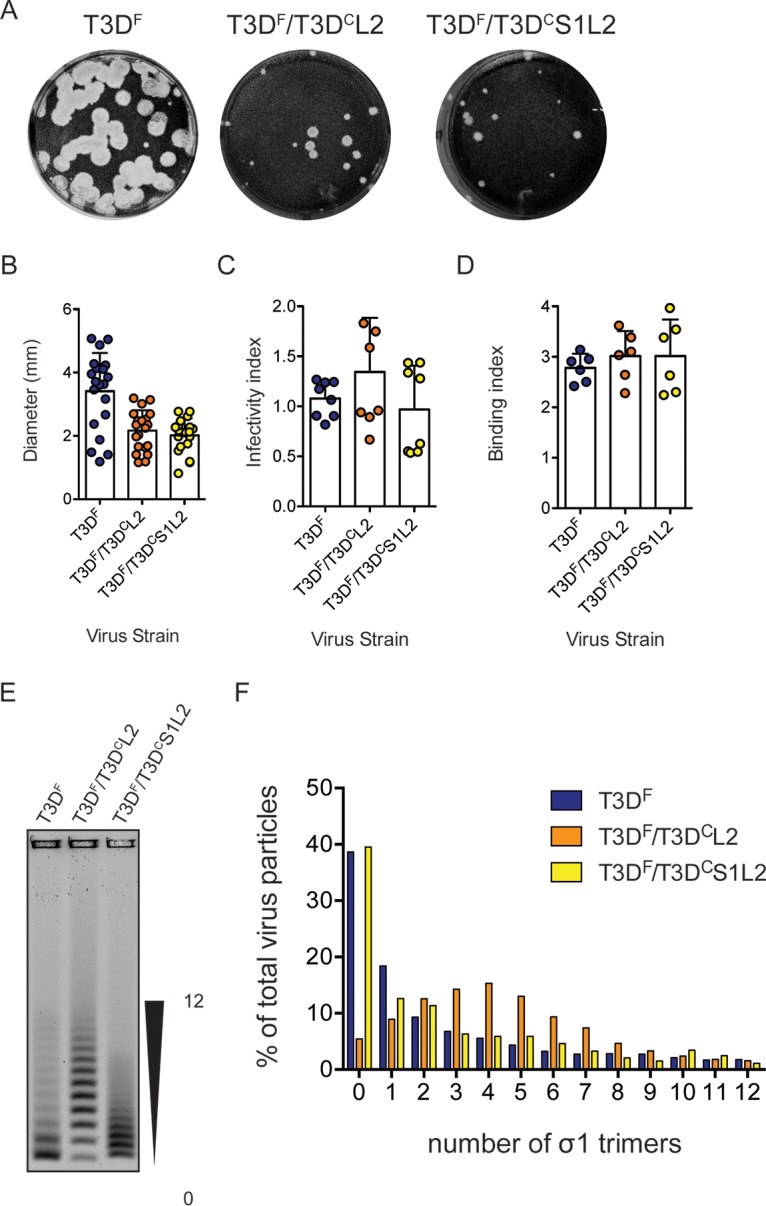
Matched L2 gene from T3DC restores σ1 encapsidation defect in T3DF/T3DCS1. (A) Particles of either T3DF, T3DF/T3DCL2, or T3DF/T3DCS1L2 diluted in PBS were subjected to plaque assay on L929 cells. This plaque assay was performed in parallel with one shown in Fig. 1. (B) Size of 20 randomly selected plaques and means are shown. Error bars indicate SD. *, P < 0.05 as determined by Student's t test compared to T3DF. The same data for T3DF as that shown in Fig. 1 were used. (C) L929 cells were adsorbed with 3,000 particles/cell of either T3DF, T3DF/T3DCL2, or T3DF/T3DCS1L2. After incubation at 37°C for 18 h, the cells were subjected to indirect immunofluorescence assay using a LI-COR Odyssey scanner. Relative infectivity was determined by calculating intensity ratios at 800 nm (green fluorescence), representing viral antigen, and 700 nm (red fluorescence), representing the cell monolayer. The infectivity index for each independent infection and the sample means are shown. Error bars indicate SD. NS, P > 0.05 as determined by 1-way ANOVA with Bonferroni's multiple-comparison test compared to T3DF. (D) Confluent monolayers of L929 cells grown in 96-well plates were adsorbed with 5 × 104 particles/cell of either T3DF, T3DF/T3DCL2, or T3DF/T3DCS1L2 at 4°C for 1 h. Cell attachment was determined by indirect immunofluorescence of cell-associated particles using a LI-COR Odyssey scanner. The binding index was determined by calculating intensity ratios at 800 nm (green fluorescence), representing viral antigen, and 700 nm (red fluorescence), representing the cell monolayer. Binding index for each independent infection and the sample mean are shown. Error bars indicate SD. NS, P > 0.05 as determined by 1-way ANOVA with Bonferroni's multiple-comparison test compared to T3DF. (E and F) Virions (1 × 1011 particles) were resolved on an agarose gel, stained with a colloidal blue staining kit, and scanned using a LI-COR Odyssey scanner. (E) The position of virions with the lowest and highest numbers of σ1 trimers is shown. (F) The band intensity of each virion species was quantified. The abundance of each species as a percentage of all 13 virion species present in the sample is shown.
