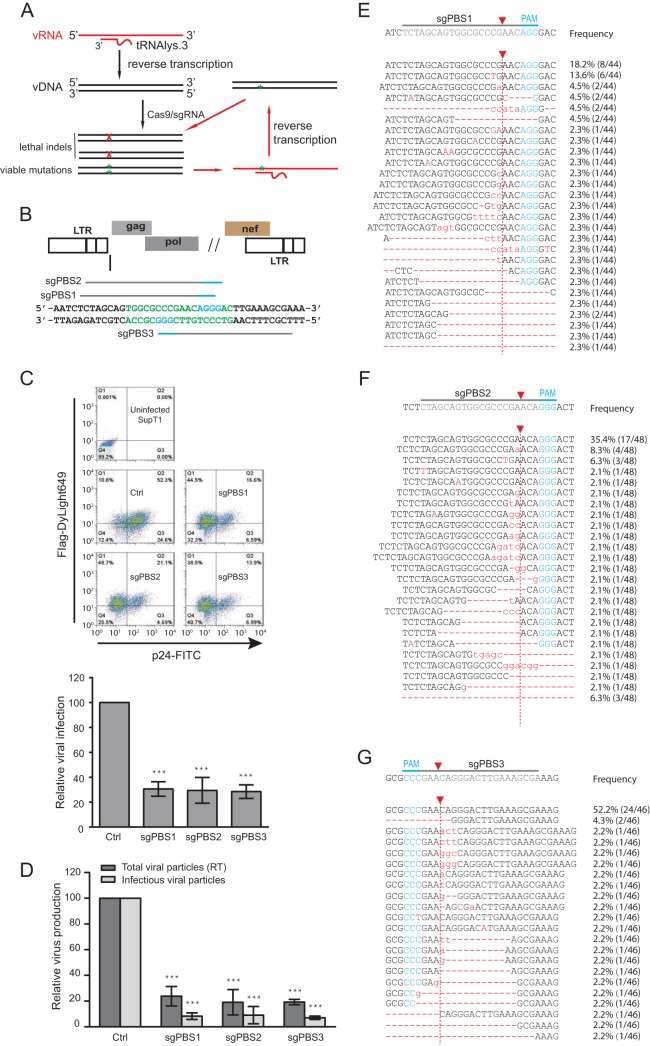
FIG 1.
Inhibition of HIV-1 infection by Cas9/sgRNA targeting viral PBS. (A) Illustration of the potential mechanisms by which the PBS-targeting Cas9/sgRNA inhibits HIV-1 infection. When the viral RNA carrying the resistant mutation in the PBS is reverse transcribed into viral DNA, the plus strand has the wild-type PBS sequence, thus potentially becoming susceptible to cleavage by Cas9/sgRNA again. vRNA, viral RNA; vDNA, viral DNA. (B) Location of the PBS-targeting sgRNA. The PBS sequence is highlighted in green letters. The PAM is indicated with blue lines and letters. (C) HIV-1 strain NL4-3 was used to infect SupT1 cells that stably express Cas9 and PBS-targeting sgRNA. Expression of Cas9 and viral p24 was examined by immunostaining and flow cytometry. Results of one representative infection experiment are shown. Results of three independent experiments are summarized in the bar graph, with the values of the control set at 100. ***, P < 0.001. (D) Levels of HIV-1 in the supernatants of infected SupT1 cells were determined either by measuring viral reverse transcriptase activity (representing total viral particles) or by infecting the TZM-bl indicator cells (representing infectious viral particles). Results of three independent infection experiments were averaged and are shown in the bar graph, with values of the control set at 100. ***, P < 0.001. (E, F, G) Indels caused by the PBS-targeting Cas9/sgRNA. Total cellular DNA was extracted from the infected SupT1 cells that expressed the sgPBS1, sgPBS2, or sgPBS3 guide RNA. Viral DNA was amplified by PCR and sequenced. Indels are shown by red letters for sgPBS1 (E), sgPBS2 (F), and sgPBS3 (G). The PAM is highlighted in blue letters. Red arrow indicates Cas9 cleavage site. The frequency of each indel among the sequenced DNA clones is presented. Error bars show standard deviations.