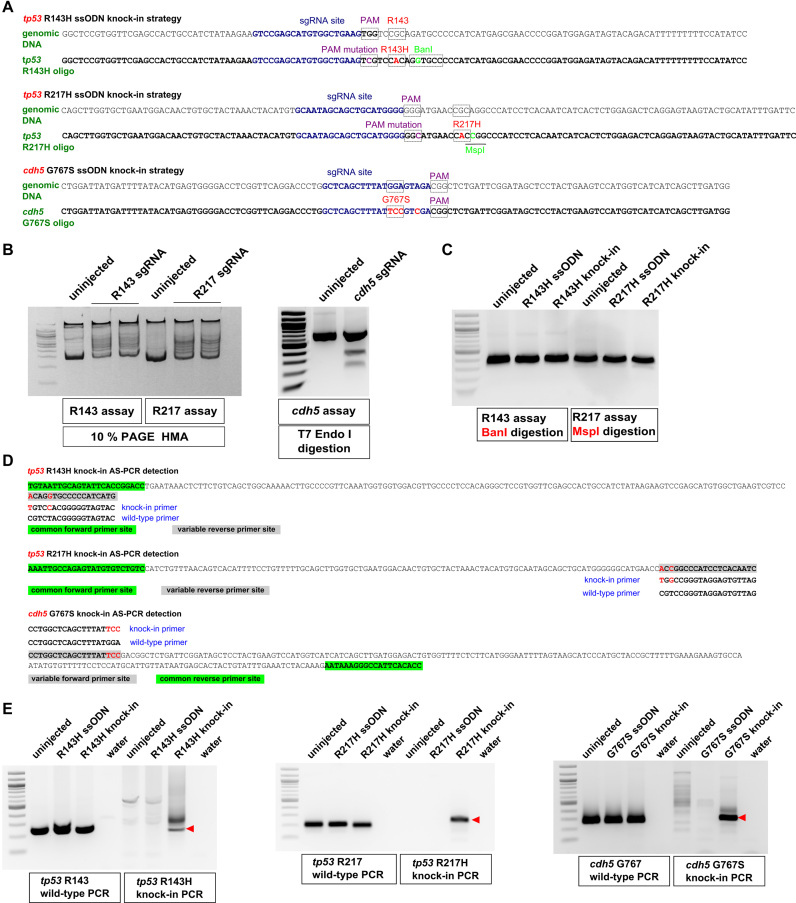
Figure 1.
Design of knock-in strategies for tp53 and cdh5 point mutations and the AS-PCR assays for detection. (A) Genomic sequences and donor oligos are shown for sites of tp53 R143H, R217H and cdh5 G767S knock-ins. sgRNA sites are shown in dark blue, PAM sites and target codons are boxed and their mutations are highlighted in magenta and red, respectively. Mutations leading to introduction of BanI (boxed) and MspI (underlined) restriction sites are highlighted in light green. (B) Mutations induced by tp53 sgRNAs were detected by HMA using 10% PAGE analysis. Detection of indel mutations induced by cdh5 sgRNA was performed using the T7 Endonuclease I assay. Comparison of PCR product samples from uninjected zebrafish embryos with those injected with respective sgRNAs shows the degree of sgRNA effectiveness. (C) Restriction analysis of PCR products from uninjected, oligo-injected and tp53 knock-in injected embryos with enzymes introduced into donor oligos. (D) Allele-specific PCR assays for detecting introduced knock-ins are based on the idea that when two or more nucleotides are different between the WT and knock-in alleles, it is possible to design primers that can distinguish the two. Primers common to both WT and knock-in assays are highlighted in green and the site for variable primers is highlighted in gray. The WT and knock-in primers are indicated below or above the site and the nucleotides corresponding to the knock-in allele are in red both in the amplicon and the knock-in primer. (E) Example AS-PCRs are shown that were run on the extracts of pooled embryos from uninjected, oligo-injected, knock-in-injected samples and water. Both WT and knock-in AS-PCRs are shown, with the WT PCRs typically being very strong and indicative of the expected size for the correct knock-in AS-PCR products (indicated with red arrowheads).