Figure 4.
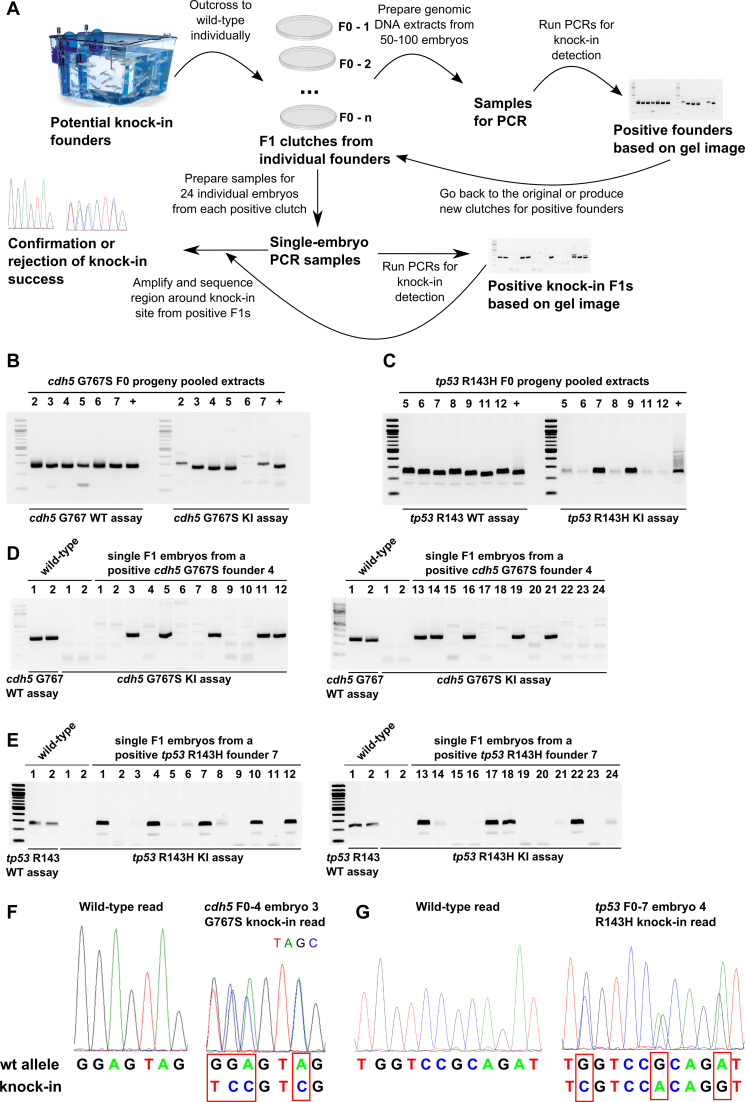
AS-PCR-based strategy enables rapid screening and confirmation of potential founders and knock-in F1 embryos. (A) In the first step of this workflow, fish injected with the verified knock-in mix (Cas9, sgRNA and oligo) are grown to adulthood and then outcrossed to WT fish. The clutches derived from the breedings are used to prepare pooled genomic DNA samples from 50–100 embryos. WT and knock-in PCR assays are then run on these samples to identify the founders with detectable levels of germ-line transmission and to provide the size marker for knock-in AS-PCR products, which should be the same size as WT assay products. The information from the first screening round is then used to determine which founders should be bred or which available clutches should be chosen for subsequent screening of 24 individual embryos from each clutch. Knock-in assays are then performed on single-embryo samples and upon obtaining the results, positive embryo extracts are used to amplify a region of DNA surrounding the knock-in site and the resulting PCR products are sent for sequencing to determine if correct knock-in has actually occurred. (B andC) WT and knock-in AS-PCR are shown for cdh5 G767S (B) and tp53 R143H (C) knock-in screening of extracts from embryo clutches produced by potential founders. (D andE) Screening examples of 24 individual embryos from cdh5 G767S and tp53 R143H knock-in founders by AS-PCR. WT and knock-in AS-PCR on two WT embryo samples are shown as controls for PCR product size and as negative controls, respectively. (F) Sequencing chromatograms from WT and heterozygous cdh5 G767S F1 embryos and alignment of the corresponding sequencing reads confirm successful knock-in at the genomic level. (G) Sequencing chromatograms from WT and heterozygous tp53 R143H F1 embryos and alignment of the corresponding sequencing reads confirm successful knock-in at the genomic level.