Abstract
MicroRNAs (miRNAs) are a class of small non-coding RNAs that repress gene expression. In plants, the RNase III enzyme Dicer-like (DCL1) processes primary miRNAs (pri-miRNAs) into miRNAs. Here, we show that SMALL1 (SMA1), a homolog of the DEAD-box pre-mRNA splicing factor Prp28, plays essential roles in miRNA biogenesis in Arabidopsis. A hypomorphic sma1-1 mutation causes growth defects and reduces miRNA accumulation correlated with increased target transcript levels. SMA1 interacts with the DCL1 complex and positively influences pri-miRNA processing. Moreover, SMA1 binds the promoter region of genes encoding pri-miRNAs (MIRs) and is required for MIR transcription. Furthermore, SMA1 also enhances the abundance of the DCL1 protein levels through promoting the splicing of the DCL1 pre-mRNAs. Collectively, our data provide new insights into the function of SMA1/Prp28 in regulating miRNA abundance in plants.
INTRODUCTION
In plants, miRNAs are involved in various biological processes, such as growth and developmental regulation, abiotic and biotic stress responses, by targeting mRNA transcripts for degradation or translational repression (1–4). The biogenesis of most plant miRNAs begins with the transcription of primary miRNAs (pri-miRNAs) by the RNA polymerase II (Pol II) from miRNA encoding genes (MIRs) (5). Following transcription, pri-miRNAs are processed by the DICER-LIKE 1 (DCL1) complex, which is composed of the RNase III enzyme DCL1, the Zinc-finger protein SERRATE (SE) and the double-stranded RNA (dsRNA)-binding protein HYPONASTIC LEAVES 1 (HYL1), to release the miRNA/miRNA* duplexes in the nucleus (2,6–8). Among DCL1 components, DCL1 directly cleaves pri-miRNAs while SE and HYL1 improve the efficiency and accuracy of DCL1 activity (9–11). After processing, miRNAs are methylated by the miRNA methyltransferase HUA ENHANCER 1 (HEN1) to prevent untemplated uridine addition that leads to degradation (12–14), and then are loaded into the effector protein named ARGONAUTE 1 (AGO1) to recognize target mRNA transcripts for cleavage or translational inhibition (15–18).
Recently, many proteins have been shown to act in miRNA biogenesis pathway. At the transcriptional level, the transcriptional co-activator Mediator (19), NEGATIVE ON TATA LESS 2 (NOT2) (20), CELL DIVISION CYCLE 5 (CDC5) (21), Elongator (22) and CYCLIN-DEPENDENT KINASES (CDKs) (23) modulate the transcriptional activity of Pol II. After transcription, the RNA-binding proteins DAWDLE (DDL) (24) and PLEIOTROPIC REGULATORY LOCUS 1 (PRL1) (25) stabilize pri-miRNA transcripts, and then the RNA-binding protein TOUGH (TGH) (26), the THO/TREX complex (27) and the ribosomal protein STV1 (28) promote the recruitment of pri-miRNAs to the DCL1 complex. At the pri-miRNA processing step, the MOS4-associated complex (MAC), NOT2, Elongator, DDL and TGH act as facilitators of the DCL1 activity (20–22,24–26,29). The activity of the DCL1 complex is also subjected to post-translational regulation. For instance, the phosphorylation status (30–32) and transportation from cytoplasm to nucleus of HYL1 protein is crucial for pri-miRNA processing (30,33). Moreover, the proper level of DCL1 can be transcriptionally or post-transcriptionally regulated by the splicing factor STABILIZED 1 (STA1) (34) and the transcription factor XAP5 CIRCADIAN TIMEKEEPER (CMA33/XCT) (35). Interestingly, miRNA biogenesis and pre-mRNA splicing share some protein factors, such as the MAC complex, STA1 (34), CAP-BINDING PROTEINs (CBPs) (36) and SE (37), suggesting the potential interplay between miRNA biogenesis and mRNA metabolism.
Prp28 is a conserved spliceosomal protein in eukaryotes. It encodes a DEAD-box helicase and is a component of U4/U5/U6 tri-small nuclear ribonucleoprotein (snRNP) in yeast and human. Prp28 is required for spliceosome assembly and functions in an adenosine triphosphate (ATP)-independent manner (38–41). Dysfunction of Prp28 in yeast and animals results in disorders of cell proliferation and differentiation (42). Compared with other organisms, plant Prp28 ortholog remains to be identified.
In this study, we report that the Prp28 homolog SMALL 1 (SMA1) contributes to miRNA biogenesis in Arabidopsis. A partial loss-of-function sma1-1 mutant displays severe growth defects, while a null sma1-2 mutant is embryonic lethal. In sma1-1, the accumulation of miRNAs and pri-miRNAs is reduced, suggesting that SMA1 is required for miRNA biogenesis. SMA1 interacts with the DCL1 complex and influences pri-miRNA processing. In addition, SMA1 is crucial for proper splicing of the DCL1 pre-mRNAs and pri-miRNAs. Moreover, SMA1 binds the MIR promoters and promotes MIR transcription through enhancing the occupancy of Pol II at the MIR promoters. Taken together, these results demonstrated that SMA1 plays a multifaceted role in miRNA biogenesis and revealed a new function for a conserved protein.
MATERIALS AND METHODS
Plant materials and growth condition
All plant materials used in this study are in the Columbia genetic background (Col-0). sma1-1 contains a G-to-T mutation at the nucleotide position 1900-bp, which can be identified through NdeI digestion of polymerase chain reaction (PCR) product amplified with the primer pairs of 730-F and 730-R. sma1-2 (SALK_110742) was obtained from the Nottingham Arabidopsis stork Center (NASC). sma1-sgR lines were generated by CRISPR/Cas9-induced mutation. Transgenic line containing a single copied pMIR167a::GUS or p35S::HYL1-YFP were crossed to sma1-1. In the F2 generation, WT plants or sma1-1 harboring pMIR167a::GUS or p35S::HYL1-YFP were selected through PCR-based genotyping for sma1-1, GUS or GFP. Approximately 15 WT or sma1-1 plants were pooled for GUS transcript level analyses. The primers used for genotyping are listed in Supplementary Table S1.
To germinate plants, seeds were sterilized using 50% bleach for 10 min, followed by four-time washes with sterile distilled water. After vernalization at 4°C for 3 days, seeds were sown on medium containing 1/2 Murashige and Skoog (MS), 1% glucose and 0.8% agar. 10-day-old seedlings were transferred to soil. All plants were grown at 22°C with 16-h light and 8-h dark cycles in a growth chamber.
Map-based cloning of SMA1
To map the sma1-1 mutation, sma1-1 was crossed with Landsberg ereca (Ler) to generate F2 population. Map-based cloning were performed with the molecular markers listed in the Supplementary Table S1. sma1-1 mutation was mapped into a 31-kb interval between markers T1B8-9 and T1B8-12 on the chromosome 2. The sma1-1 mutation within this region was then identified through DNA-sequencing.
Plasmid construction
The protein encoding region of SMA1 was PCR-amplified with primers SMA1CDS-F and SMA1CDS-R, cloned into PCR8/GW/TOPO cloning vector (Invitrogen) and subsequently subcloned into pMDC43 binary vector to generate 35S::GFP-SMA1 construct. To generate the bimolecular fluorescence complementation (BiFC) construct for SMA1, SMA1 cDNA was inserted into the multiple clone sites of pSAT4-cCFP-C vector. Then, the resulting plasmid was digested with the I-SceI restriction enzyme to release the 35S::cCFP-SMA1 fragment, which was then subcloned to pPZP-RCS2-ocs-bar-RI vector. The constructs of nVenus-DCL1, nVenus-SE, nVenus-HYL1 and nVenus-AGO1 were described previously (26). The construct for CRISPR/Cas9-induced SMA1 mutation was generated with binary vector pHEE401 using BsaI (NEB) and T4 ligase (NEB) according to the method described previously (43). The primers were listed in Supplementary Table S1.
Morphological analyses and GUS histochemical staining
Morphological and cellular analyses were performed according to the previously reported methods (44). For GUS staining, tissues from sma1-1 or WT plants harboring pMIR167a::GUS were incubated for 5 h in the staining solution (1 mM 5-bromo-4-chloro-3-indoly-β-D-glucuronic acid, 100 mM NaPO4 buffer, 3 mM each K3Fe(CN)6/K4Fe(CN)6, 10 mM EDTA and 0.1% NP-40) at 37°C. Seventy percent ethanol was used for tissue clearing before imaging.
Detection of DCL1, SE, HYL1 and AGO proteins in Arabidopsis
Protein extracts from inflorescences of Arabidopsis were separated on a 10% Sodium dodecylsulphate-polyacrylamide gel electrophoresis (SDS-PAGE) and detected with antibodies against HYL1 (Agrisera, AS06136), SE (Agrisera, AS09532A), DCL1 (Agrisera, AS122102), AGO1 (Agrisera, AS09527) and HSC70 (Enzo, ADI-SPA-818).
BiFC assay
BiFC assay was performed as described (21). The constructs of cCFP-SMA1, nVENUS-DCL1, nVENUS-SE, nVENUS-HYL1 and nVENUS-AGO1 were transformed into Agrobacterium strains. All these transgenes are under the control of the cauliflower mosaic virus 35S promoter (35S promoter). The transformed Agrobacterium strains were then collected and suspended in infiltration buffer (10 mM MES pH 5.6, 150 mM acetosyringone and 10 mM MgCl2). Paired C-terminal fragment of cyan fluorescent protein (cCFP) and nVENUS fusion proteins were co-expressed in Nicotiana benthamiana leaves. After 40 h expression, a confocal microscopy (Fluoview 500 workstation; Olympus) was used to detect YFP and chlorophyll auto fluorescence signals at 488 nm with a narrow barrier (505–525 nm, BA505–525; Olympus).
Co-IP assay
To examine the interaction of SMA1 with DCL1 and SE, GFP-SMA1 was transiently co-expressed with MYC-DCL1 or MYC-SE in N. benthamiana as described (26). The expression of these transgenes was directed by the 35S promoter. Total protein was extracted with extraction buffer (50 mM Tris–HCl 8.0, 150 mM NaCl, 5% glycerol, 5% Triton X-100, 1 mM EDTA, 1× complete protease inhibitor cocktail and 1 mM phenylmethylsulfonyl fluoride). Immunoprecipitation (IP) was performed on protein extracts using anti-GFP antibodies (GTA020, Bulldog Bio) coupled to protein G agarose beads. After IP, proteins were separated on a 10% SDS-PAGE and detected with western blot using monoclonal antibodies against GFP (Biolegend, 902602) or MYC (Millipore, 06–340).
ChIP assay
Chromatin immunoprecipitation assay (ChIP) was performed using 14-d-old seedlings of Col-0, sma1-1 and sma1-1 containing 35S::GFP or 35S::GFP-SMA1 as described (19). Three biological replicates were performed. Antibodies of C-terminal domain (CTD) (ab817, Abcam), the Ser5P form of Pol II CTD (ab5408, Abcam), the Ser2P form of Pol II CTD (ab5095, Abcam) or GFP (GTA020, Bulldog Bio) were used for IP. Quantitative PCR (qPCR) was performed using primers listed in Supplementary Table S1.
RNA isolation, northern blot and qRT-PCR analyses
Total RNA was extracted with Trizol reagent. Northern blot was performed as described (26). Approximately 15 μg total RNAs extracted from inflorescences were resolved on 16% PAGE gel and transferred to nylon membranes. 32P-labeled antisense DNA oligonucleotides were used to detect small RNAs. Radioactive signals were detected with a Phosphorimager and quantified with ImageQuant.
The levels of pri-miRNAs, miRNA target transcripts and GUS mRNA were determined using qRT-PCR. A total of 2 μg total RNAs from inflorescences were used to generate cDNAs using the SuperScript III reverse transcriptase (Invitrogen) and an oligo dT18 primer. cDNAs were then used as templates for qPCR on an iCycler apparatus (Bio-Rad) with the SYBR Green Master Mix (Bio-Rad). The primers used for PCR are listed on Supplementary Table S1.
Small RNA sequencing
Inflorescence tissues of Col-0 and sma1-1 were harvested as two biological replicates and used for small RNA sequencing. The generated sequencing data were analyzed for miRNA abundance according to Ren et al. (26). The dataset was deposited into National Center for Biotechnology Information Gene Expression Omnibus (Col-0 accession numbers: GSM3132081 and GSM3132082; sma1-1 accession numbers: GSM3132083 and GSM3132084).
RESULTS
SMA1 is required for development and miRNA accumulation
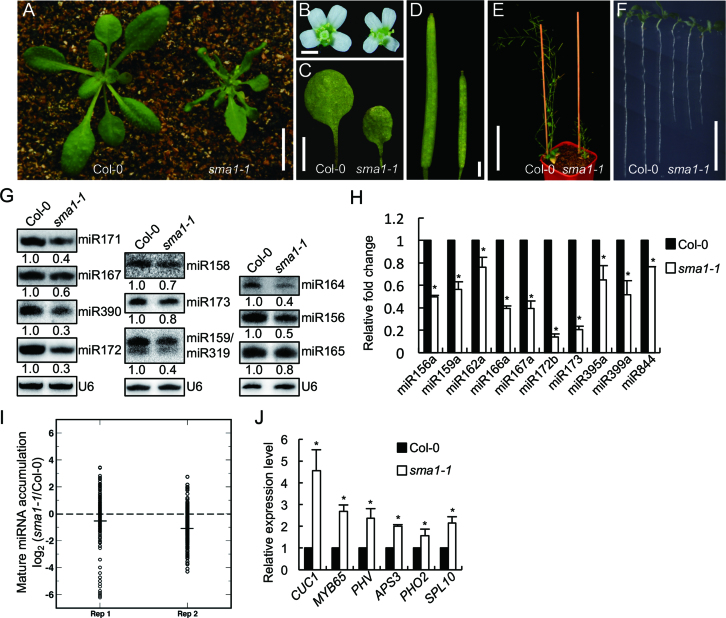
From a Col-0 (wild-type, WT) population harboring a proMED8::GUS transgene, we isolated a spontaneous mutant with tiny plant size that we named as small1-1 (sma1-1). sma1-1 was backcrossed to Col-0 for five times to remove the proMED8::GUS transgene and other potential mutations before further investigation. Compared with WT, sma1-1 displayed growth defects with reduced organ size, dwarf and short roots (Figure 1A–F). We suspected that sma1-1 may contain less amounts of miRNAs because its pleiotropic growth defects agree with the role of miRNA in controlling plant development. To test this hypothesis, we compared miRNA levels in sma1-1 with those in Col-0 by Northern blot. All 11 tested miRNAs were decreased in abundance in sma1-1 relative to Col-0 (Figure 1G). qRT-PCR analyses further confirmed that the accumulation of miRNAs was reduced in sma1-1 relative to Col-0 (Figure 1H). These results demonstrate that SMA1 is required for miRNA accumulation. To determine the effect of SMA1 on miRNA expression at global levels, we further analyzed miRNA profile from inflorescences of sma1-1 and WT through illumina deep sequencing. Many miRNAs were reduced in abundance in sma1-1 relative to WT (Figure 1I and Supplementary dataset 1), revealing that SMA1 has a general effect on miRNA accumulation. We also examined the effect of sma1-1 on the accurate cleavage of pri-miRNAs, meaning that sequenced miRNAs or miRNA* should fall within ±2 bases of the annotated mature miRNA(s) or miRNA*(s) positions (26). The ratio of imprecise miRNAs in sma1-1 was similar with that of WT (Supplementary dataset 2), suggesting that SMA1 did not affect the accurate cleavage of pri-miRNAs.
Figure 1.
sma1-1 causes pleiotropic developmental defects and reduces the accumulation of miRNAs. (A) Four-week-old plants of Col-0 and sma1-1. (B) Opening flowers of Col-0 and sma1-1 (from left to right). (C) The first-pair of leaves of Col-0 and sma1-1. (D) The mature siliques of Col-0 and sma1-1 (from left to right). (E) Seven-week-old plants of Col-0 and sma1-1. (F) Seven-day-old seedlings of Col-0 and sma1-1. (G) The accumulation of miRNAs in Col-0 and sma1-1 detected by northern blot. The U6 RNA was shown as a loading control. The numbers below the picture indicate the relative amount of small RNAs in sma1-1 and Col-0 and represent mean of three replicates (P < 0.05). In the miR159/miR319 blot, the upper band is miR159 whereas the lower band represents miR319. (H) The levels of miRNAs detected by RT-qPCR. miRNA levels in sma1-1 were normalized to those of U6 RNAs and compared with Col-0 (set as 1). (I) Small RNA sequencing analysis of miRNAs in sma1-1 and Col-0. The miRNA abundance was calculated as reads per million, and a log2-transformed ratio of sma1-1/Col-0 was plotted. Each circle represents one miRNA. Think lines indicate median values. (J) The levels of miRNA target transcripts in Col-0 and sma1-1 detected by RT-qPCR. The levels of miRNA target transcripts were normalized to those of UBIQUITIN 5 (UBQ5) and compared with Col-0 (set as 1). Error bars in (H) and (J): standard deviations (SD) of three replicates (*: P < 0.05 with one-sample t-test). Scale bars: 1 cm (A), 1 mm (B), 5 mm (C), 1 mm (D), 10 cm (E) and 1 cm (F).
We next tested the effect of SMA1 on the accumulation of miRNA target transcripts, since miRNAs mainly repress the transcript levels of their targets. qRT-PCR analyses showed that relative to Col-0, sma1-1 increased the levels of several examined target transcripts, including CUC1, MYB65, PHV, APS3, PHO2 and SPL10, which are targets of miR164, miR159, miR166, miR395, miR399 and miR156, respectively (Figure 1J). This result is consistent with the decreased levels of miRNAs in sma1-1.
SMA1 encodes a pre-mRNA splicing factor
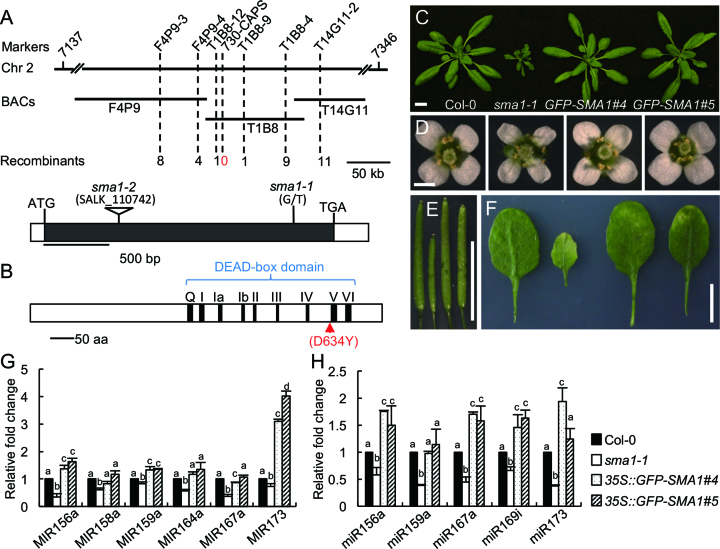
To identify how SMA1 affects miRNA accumulation, we used a map-based cloning approach to identify the sma1-1 mutation. The mutation was mapped to a 31-kb region of chromosome 2 (Figure 2A). DNA sequencing analysis of this region revealed a G-to-T transversion at nucleotide position 1900 in the AT2G33730 gene (Figure 2A). To confirm if this mutation is responsible for the observed phenotypes of sma1-1, the protein-encoding region of AT2G33730 was cloned into pMDC43 vector to generate 35S::GFP-SMA1 construct and was introduced into sma1-1. The expression of the 35S::GFP-SMA1 transgene rescued the growth defects in the sma1-1 mutant, including plant size and height (Figure 2C–E and Supplementary Figure S1). Moreover, in transgenic plants, the levels of miRNA and pri-miRNA were comparable to those in Col (Figure 2G and H)). These results demonstrate that AT2G33730 indeed encodes SMA1.
Figure 2.
Identification of SMA1 gene and complementation of sma1-1. (A) Map-based cloning of the sma1-1 mutation and diagram of the SMA1 gene structure. The sma1-1 mutation was mapped to the region between markers T1B8-12 and T1B8-9 on the chromosome 2, and co-segregated with marker 730-CAPS. ATG: the start codon. TGA: the stop codon. Black box: the coding sequence. White boxes: the 5′ or 3′ untranslated regions. The mutation sites of sma1-1 and sma1-2 (SALK_110742) are shown above the SMA1 gene structure. (B) The predicted domains of the SMA1 protein. The nine conserved motifs in the DEAD-box domain are indicated. The G-to-T mutation in sma1-1 causes a D (Asp)-to-Y (Tyr) change at the amino acid position 634. (C–F) The 35S::GFP-SMA1 transgene rescues the growth defects of sma1-1 in 4-week-old seedlings (C), opening flowers (D), mature siliques (E) and the fifth leaves (F). Scale bars: 1 cm (C), 1 mm (D) and 1 cm (E and F). (G) and (H) The levels of pri-miRNAs (H) and mature miRNAs (H) in 35S::GFP-SMA1 transgenic plants were detected by RT-qPCR. The levels of pri-miRNAs and miRNAs were normalized to UBQ5 and U6, respectively. The numbers represent mean of three replicates relative to Col-0 (set as 1). Error bars in (G) and (H): standard deviations (SD) of three replicates. Means with different letters are significantly different determined by ANOVA (P < 0.05).
SMA1 is a conserved protein in plants (Supplementary Figure S2). It encodes a peptide of 733 amino acids, which shares considerable similarity (42%) with yeast pre-mRNA splicing factor Prp28 (Supplementary Figure S3). Like Prp28, SMA1 contains a putative DEAD-box domain with nine conserved motifs, which are required for ATPase and RNA unwinding activity (Figure 2B and Supplementary Figure S3) (45). The sma1-1 mutation caused the conversion of an (conserved/invariant) Aspartic acid (D) in the conserved DEAD-box domain to Tyrosine (Y) at the amino acid position 634 (Figure 2B and Supplementary Figure S3).
We also obtained a second sma1 allele (SALK_110742, sma1-2) with a T-DNA insertion (Supplementary Figure S4B). However, no homozygous sma1-2 plant was identified from self-crossed population of sma1-2/+. Moreover, the immature siliques from heterozygous plants contained ∼25% of aborted seeds (Supplementary Figure S4C), suggesting the arrest of embryo development. We crossed sma1-1 with the heterozygous plants of sma1-2. The F1 plants with sma1-1/+ genotype developed normally as the WT, while F1 plants with sma1-1/sma1-2 genotype exhibited the sma1-1 phenotypes (Supplementary Figure S4D). These results reveal that sma1-2 is likely a null mutation and causes embryo lethality when becoming homozygous. We next used a CRISPR/Cas9 system to generate two additional alleles of sma1, smal1-3 and smal1-4, which contain a nucleotide insertion and a deletion of 32 bp nucleotides at the 3′ end of the SMA1 gene, respectively (Supplementary Figure S5A). Compared with WT, these two mutants were defective in development and displayed reduced miRNA abundance (Supplementary Figure S5C), which further support that SMA1 functions in miRNA biogenesis.
SMA1 regulates DCL1 pre-mRNA splicing
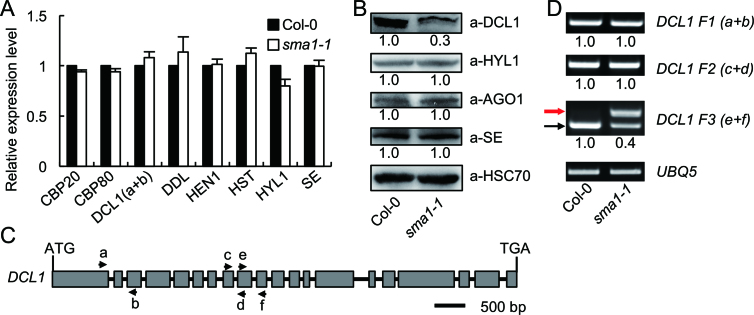
Because SMA1 is a Prp28 homolog, we reasoned that it might function as a splicing factor to control the expression levels of genes involved in miRNA biogenesis. We therefore examined the transcription levels of the well-known key factors involved in miRNA biogenesis including CBP20, CBP80, DCL1, DDL, HEN1, HST, HYL1 and SE through qRT-PCR. The transcript levels of these genes were indistinguishable between Col-0 and sma1-1 mutant (Figure 3A). To confirm the result, we examined the protein levels by western blot using the antibodies against DCL1, HYL1, AGO1 and SE. The results showed that the protein levels of HYL1, AGO1, and SE were not changed in sma1-1 (Figure 3B). In contrast, DCL1 protein levels were decreased in the sma1-1 mutant compared with Col-0 (Figure 3B). We next tested if the splicing of DCL1 pre-mRNAs was impaired in sma1-1. Several pairs of primers spanning various introns (Figure 3C) were used to examine the splicing of the DCL1 pre-mRNA through RT-PCR analysis in sma1-1 and Col-0. Compared with Col, sma1-1 did not show obvious effect on the splicing of the firsrt, second and eighth introns of the DCL1 pre-mRNAs (Figure 3D). However, the retention of the ninth intron was increased in sma1-1 relative to Col-0 (Figure 3D), suggesting that SMA1 may regulate the splicing of DCL1 pre-mRNA, which is likely attribute to the decreased DCL1 protein levels in sma1-1. The effect of sma1-1 on the splicing of DCL1 pre-mRNA suggests that SMA1 may have a general role in splicing of protein-coding genes. To test this possibility, we examined the splicing of additional 18 pre-mRNAs including HEN1, HYL1, SE, DDL and others (Supplementary Figure S6A). The result showed that smal-1 increased intron retention of four pre-mRNAs (Supplementary Figure S6B), indicating SMA1 may have a general role in intron splicing.
Figure 3.
The expression levels of factors involved in miRNA biogenesis. (A) The transcript levels of various genes in sma1-1 and Col-0 determined by RT-qPCR. The positions of primers (a and b) used for DCL1 amplification are shown in (C). The expression levels of examined genes were normalized to UBQ5. Gene expression levels in sma1-1 represent mean of three replicates relative to Col-0 (set as 1). (B) The protein levels of DCL1, HYL1, AGO1 and SE detected by their specific antibodies in Col-0 and sma1-1. Heat Shock Protein 70 (HSC70) was employed as loading control. Three replicates were performed and one representative figure was shown for each protein. The numbers shown below the membranes indicate the normalized values relative to Col-0 (set as 1). (C) Diagram showing the gene structure and primer pairs used for DCL1 amplification. (D) Intron-retention analysis of the DCL1 pre-mRNA using indicated primer pairs. UBQ5 served as the loading control. Upper arrow indicates the unspliced transcripts. Lower arrow shows the spliced transcripts. Three replicates were performed and one representative figure was shown for each PCR amplification. The numbers shown below the figures indicate the ratio of spliced transcripts to total transcripts.
SMA1 affects pri-miRNA splicing
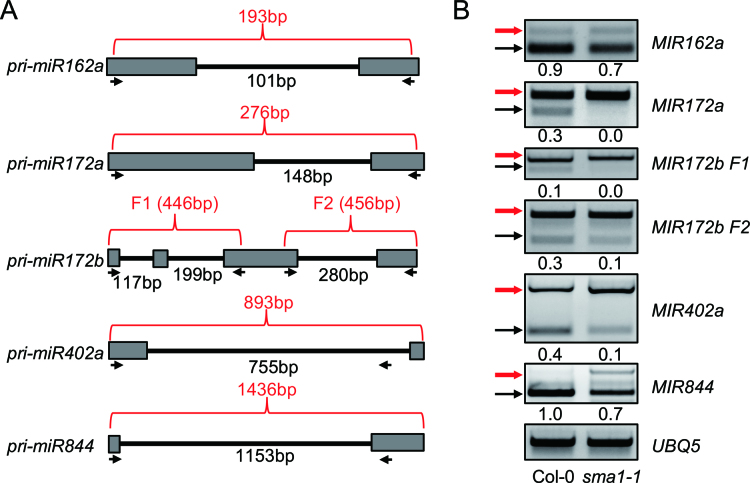
The defection of DCL1 pre-mRNA splicing in sma1-1 suggests that like Prp28 in yeast and animals, SMA1 likely participates in splicing. Thus, we sought to examine if sma1-1 affected the splicing of intron-containing pri-miRNAs through RT-PCR using primers spanning the intron region of pri-miRNAs (Figure 4A). Increased intron retention was detected in all five examined pri-miRNAs in sma1-1 compared with Col-0 (Figure 4B). This result reveals that SMA1 affects the splicing of some pri-miRNAs.
Figure 4.
Intron-retention analysis of pri-miRNA transcripts. (A) Diagram showing the partial regions of several intron-containing pri-miRNAs. Arrows indicate primers used for PCR. The numbers in red indicate the length of PCR products without intron splicing. The numbers in black show the intron length. (B) Intron-retention of pri-miRNAs analyzed by RT-PCR. Upper arrows indicate the unspliced transcripts. Lower arrows indicate the intron-spliced transcripts. UBQ5 served as the loading control. Three replicates were performed and one representative figure was shown for each PCR amplification. The numbers shown below the figures indicate the ratio of spliced transcripts to total transcripts.
SMA1 promotes pri-miRNA transcription
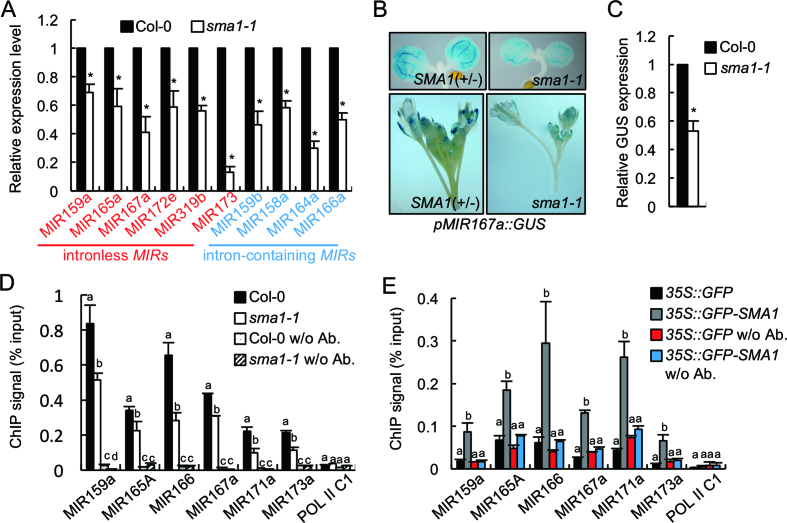
Because DCL1 is the enzyme processing pri-miRNAs, we reasoned that reduced DCL1 protein levels may impair pri-miRNA processing, resulting in increased pri-miRNA levels. To test this hypothesis, we examined pri-miRNA levels in sma1-1 and Col-0. However, qRT-PCR analyses showed that the levels of eight examined pri-miRNAs were decreased in sma1-1 relative to Col (Figure 5A). This result suggests that SMA1 may positively regulate pri-miRNA accumulation. Given the fact that the abundance of intronless pri-miRNAs such as pri-miR159a, pri-miR165a, pri-miR167a, pri-miR172e, pri-miR173 and pri-miR319b was also reduced in sma1-1 (Figure 5A), we suspected that SMA1 might have other roles in promoting pri-miRNA accumulation besides splicing. Because pri-miRNA levels are partially determined by Pol II-dependent transcription, we hypothesized that SMA1 may positively regulate MIR transcription. We used a pMIR167a::GUS reporter line, which has been used to evaluate the function of several genes in controlling MIR transcription (19,25,29), to test this hypothesis. We crossed the pMIR167a::GUS reporter line to sma1-1. In F2 population, WT plants (SMA1/SMA1, SMA1/sma1-1) and sma1-1 containing the pMIR167a::GUS transgene were selected to examine GUS expression. GUS staining of 5-day-old seedlings and inflorescences showed that GUS protein levels were decreased in sma1-1 background compared with those in WT (Figure 5B). RT-PCR analysis of RNAs isolated from inflorescences further confirmed that the GUS transcript levels were reduced in sma1-1 (Figure 5C). These results indicate that SMA1 may play a role in promoting MIR transcription. To confirm that sma1-1 indeed affects MIR transcription, we examined the occupancy of Pol II at the MIR promoters in sma1-1 and Col through the ChIP using an antibody. After IP using antibodies recognizing Ser 5-phosphorylated or unphosphorylated CTD of the largest subunit of Pol II (RPB1), qPCR analyses showed the occupancy of Pol II at the promoters of five examined MIRs was reduced in sma1-1 relative to Col-0 (Figure 5D and Supplementary Figure S7A). Because the occupancy of RPB1 with Ser 5-phosphorylated CTD at Pol II-dependent genes is positively correlated with transcription initiation (46), and the amount of RPB1 with unphosphorylated CTD indicates the recruitment of the transcription preinitiation complex to the Pol II-dependent genes (46), these results reveal that SMA1 is required for transcription initiation. We also tested Pol II occupancy the MIR genes using antibody recognizing Ser 2-phosphorylated RPB1 and found that the occupancy of Ser 2-phosphorylated RPB1 at MIR genes was increased in sma1-1 relative to WT (Supplementary Figure S7B). Because the occupancy of RBP1 with Ser 2-phosphorylated CTD is positively correlated with transcription elongation, our result suggests that SMA1 may promote elongation. However, transcription elongation is not a rate- limiting step in plants due to NELF homologs, which negatively regulate Pol II elongation, and whose effect is released by phosphorylation of Ser 2, are lacking in plants (47,48). Thus, our results suggest that SMA1 may promote transcription by modulating transcription initiation.
Figure 5.
SMA1 is required for MIR transcription. (A) Pri-miRNA levels in Col-0 and sma1-1 detected by RT-qPCR. Pri-miRNA levels in sma1-1 were normalized to those of UBQ5 and compared with Col-0 (set as 1). (B) Histochemical GUS staining of five-day-old seedlings and inflorescences in Col-0 and sma1-1 harboring the pMIR167a::GUS transgene. Fifteen plants containing GUS were analyzed for each genotype and one image for each genotype is shown. (C) GUS transcript levels in Col-0 and sma1-1 harboring the pMIR167a::GUS transgene. GUS transcript levels detected by RT-qPCR were normalized to those of UBQ5 and compared with Col-0 (set as 1). (D) The occupancy of Pol II at MIR promoters in Col-0 and sma1-1 detected by ChIP followed by qPCR. The intergenic region between At2g17470 and At2g17460 (POL II C1) was used as a negative control. IP was performed using anti-RPB1 antibodies. (E) The association of GFP and GFP-SMA1 with MIR promoters in transgenic plants containing 35S::GFP or 35S::GFP-SMA1. The occupancy of GFP-SMA1 and GFP at MIR promoters was detected by qPCR following ChIP. The intergenic region between At2g17470 and At2g17460 (POL II C1) was used as a negative control. IP was performed using anti-GFP antibodies. Error bars (A–E) indicate standard deviations (SD) of three replicates. * in A and C: P < 0.05 with one-sample t-test. Means with different letters are significantly different determined by ANOVA (P < 0.05).
Next, we asked if SMA1 could bind the MIR promoter region, which could indicate a direct role of SMA1 in regulating MIR transcription. We performed ChIP on the complementation line of sma1-1 harboring the 35S::GFP-SMA1 transgene (Figure 2) using antibodies against GFP. Indeed, qPCR detected the presence of several examined MIR promoters in GFP-SMA1 IPs but not in GFP control IPs or no-antibody controls (Figure 5E). Moreover, the negative control Pol II C1 that is the intergenic region between AT2G17470 and AT2G17460 was not detected in SMA1 IPs. Taken together, our results suggest that SMA1 may have a direct role in promoting MIR transcription. We also examined if SMA1 could bind protein-coding genes and found that SMA1 occupied at the promoter of one out of five examined genes (Supplementary Figure S7C), suggesting that SMA1 may also recognize some protein coding genes.
SMA1 associates with the DCL1 complex
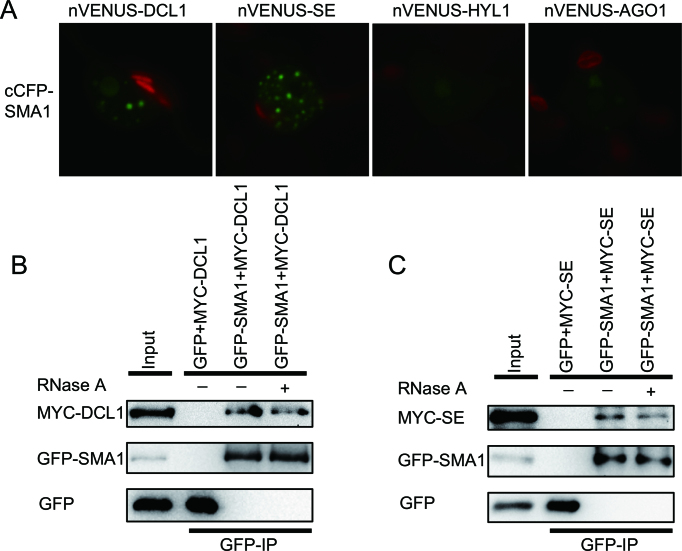
Recent studies have revealed that several splicing-related proteins such as MAC3, CDC5 and PRL1 associate with the DCL1 complex to modulate its activity (21,25,49). By analogy, we sought to test if SMA1 interacts with the DCL1 complex. We performed a BiFC assay to determine the interaction of SMA1 with DCL1, SE and HYL1. In this assay, SMA1 protein was fused with the cCFP at its C-terminus (cCFP-SMA1), while the DCL1, SE and HYL1 proteins were fused to nVENUS at their C-terminus (nVENUS-DCL1, nVENUS-SE and nVENUS-HYL1, respectively). After 2 days co-infiltration of paired proteins (cCFP-SMA1/nVENUS-DCL1, cCFP-SMA1/nVENUS-SE and cCFP-SMA1/nVENUS-HYL1), yellow fluorescence (YFP, showing in green) signals were examined with confocal microscopy. Co-expression of cCFP-SMA1 with nVENUS-DCL1 or nVENUS-SE produced strong YFP signals (Figure 6A). In contrast, the combination of cCFP-SMA1 and nVENUS-HYL1 or the negative control nVENUS-AGO1 did not generate fluorescence signal (Figure 6A). These results suggest a potential interaction of SMA1 with the DCL1 complex. We next performed a co-IP assay to confirm the SMA1-DCL1 and SMA1-SE interactions. In this experiment, we transiently co-expressed GFP-SMA1/MYC-DCL1, GFP-SMA1/MYC-SE, GFP/MYC-DCL1 or GFP/MYC-SE in N. benthamiana, and performed IP using anti-GFP antibodies. After IP, both DCL1 and SE were detected in the GFP-SMA1 precipitates but not in the GFP precipitates (Figure 6B and C). In addition, RNase A treatment did not disrupt the associations of SMA1 with DCL1 and SE (Figure 6B and C), suggesting that their interactions are independent of RNAs. The association of SMA1 with the DCL1 complex led us to test if sma1-1 affected the assembly of the DCL1 complex (D-body). We crossed a transgenic line harboring the HYL1-YFP, which has been used as a marker for D-body (20,50,51), into sma1-1 and examined the numbers of D-bodies. sma1-1 did not show obvious effect on the formation of D-bodies (Supplementary Figure S8).
Figure 6.
SMA1 associates with the DCL1 complex. (A) The interaction of SMA1 with DCL1, SE, HYL1 and AGO1 detected by BiFC. Paired cCFP- and nVenus-fusion proteins were co-expressed in Nicotiana benthamiana leaves. The combination of cCFP-SMA1 and nVENUS-AGO1 was used as a negative control. Green: BiFC signal (originally yellow fluorescence). Red: autofluorescence of chlorophyll. (B and C) Co-IP between SMA1 and DCL1 (C), SMA1 and SE (D). IP was performed using anti-GFP antibodies. Proteins of MYC-DCL1, MYC-SE, GFP-SMA1 and GFP were detected by immunoblot with anti-MYC or anti-GFP antibodies. Inputs show the total protein before IP. RNase A was used to digest RNAs.
DISCUSSION
Prp28 is a conserved protein in eukaryotes. Its roles in splicing have been documented in yeast and human (52,53). In yeast and human, PRP28 promotes the transition from a pre-catalytic spliceosome to an activated spliceosome by displacing U1 snRNP from the 5′-splice site (54,55). However, whether Prp28 has functions other than splicing is not clear. Moreover, little is known about the activity of plant Prp28 orthologs due to the lack of corresponding mutants. In this study, we find that sma1-2, a null mutant of the Arabidopsis ortholog of Prp28, is embryo lethal and that a hypomorphic sma1-1 mutant displays pleiotropic developmental defects. These results suggest that SMA1 is essential for plant development. In sma1-1, the accumulation of miRNAs is reduced. This observation together with the fact that SMA1 binds the MIR loci and interacts with the DCL1 complex suggests that SMA1 is a player in miRNA biogenesis. The impaired miRNA biogenesis shall partially contribute to the developmental defects of sma1-1. However, the roles of SMA1 in splicing and transcription shall also be important for plant development.
How does SMA1 function in miRNA biogenesis? In sma1-1, pri-miRNA accumulation is reduced. One possible explanation is that SMA1 affects pri-miRNA splicing. This opinion is evidenced by the fact that the splicing of some pri-miRNAs is impaired in sma1-1. Recent studies have suggested the crosstalk between pri-miRNA processing and splicing (56,57). Some slicing related factors are required for miRNA biogenesis (27,34,37,58–61). Splicing itself also regulates the processing of some pri-miRNAs (62–71). The observation that the accumulation of two miRNAs with defective pri-miRNA splicing is reduced in sma1-1 suggests that SMA1 may promote pri-miRNA accumulation by modulating splicing. However, SMA1 must have other roles in promoting pri-miRNA levels, because some pri-miRNAs do not show splicing defects in sma1-1 and the intronless pri-miRNAs also show reduced abundance in sma1-1. Indeed, sma1-1 impaired the MIR promoter activities (Figure 7). Moreover, SMA1 is required for Pol II occupancy at the MIR loci. These results reveal that SMA1 may function as a positive regulator of MIR transcription (Figure 7). It is not clear how SMA1 influences MIR transcription, as we did not observe the interaction of SMA1 with Pol II. Since SMA1 is a helicase, we speculate that it may function in transcription by modulating chromatin structures to facilitate the access of Pol II or other transcription factors. Consistent with this notion, SMA1 interacts with the MIR loci and the sma1-1 mutation within the helicase domain reduces MIR transcription.
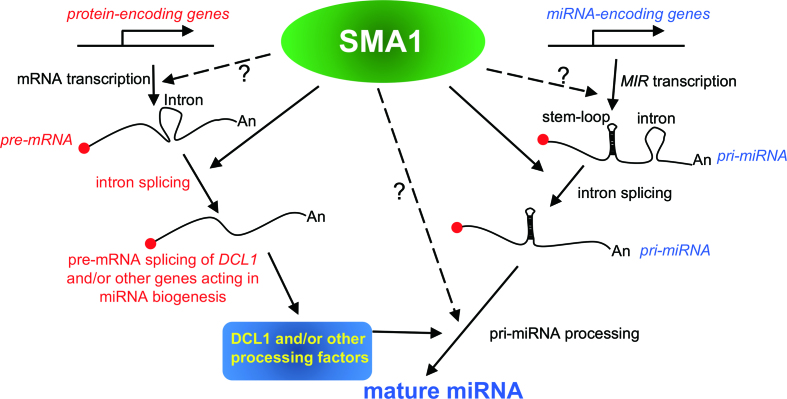
Figure 7.
A model proposing the multiple roles of SMA1 in miRNA biogenesis and mRNA metabolism. SMA1 associates with the MIR promoters to promote Pol II-dependent transcription. In addition, it can modulate the splicing of some pri-miRNAs and may function as an accessory factor of the DCL1 complex to facilitate pri-miRNA processing. Moreover, SMA1 influences pri-miRNA processing through enhancing the proper splicing of the DCL1 pre-mRNAs. SMA1 also promotes the splicing of some protein-coding genes including DCL1, and, by analogy, may regulate the transcription of some protein-coding genes.
SMA1 also enhances the accumulation of the DCL1 protein (Figure 7). The retention of the ninth intron of DCL1 pre-mRNA transcript is increased in sma1-1, suggesting that SMA1 controls DCL1 expression through promoting proper DCL1 pre-mRNA splicing (Figure 7). Since DCL1 is the enzyme catalyzing pri-miRNA processing, we propose that SMA1 indirectly influences pri-miRNA processing through modulating DCL1 protein levels. It is not clear why SMA1 only affects the splicing the ninth intron of DCL1. An explanation is that SMA1 may regulate some specific introns. Alternatively, this maybe due to that smal-1 is a weak allele.
Moreover, SMA1 associates with the DCL1 complex. In metazoans, several DEAD-box RNA helicases including p68 and p72 function as accessory components of the Drosha complex to enhance miRNA production (72–74). We envision that the interaction of SMA1 with the DCL1 complex may also facilitate pri-miRNA processing (Figure 7). Besides SMA1, several splicing related factors such as CDC5, MAC7 and MAC3A/3B also promote MIR transcription and function as accessory components of the DCL1 complex (21,29,49). Given the fact that pri-miRNAs are co-transcriptionally processed by DCL1, we suspect that a common theme for these proteins is to bridge the transcription, splicing and processing of pri-miRNAs.
In summary, we identify SMA1 as a new player in miRNA biogenesis. It positively influences miRNA accumulation through enhancing pri-miRNA transcription and/or splicing, modulating the DCL1 protein levels and facilitating the DCL1 activities (Figure 7). However, we cannot completely rule out the possibility that the observed miRNA biogenesis defects maybe caused by the developmental roadblock of sma1 that does not allow progression of the normal gene expression program. The splicing of some pri-miRNAs and some pre-mRNAs is defective in sma1-1, suggesting that SMA1 plays essential roles in splicing of a subset of Pol-II dependent RNAs. Notably, we also uncovered a role of SMA1 in MIR transcription. Since MIRs share similar gene structures with other Pol II dependent genes, we propose that SMA1 may affect the transcription of a subset of protein-coding genes. Clearly, the availability of sma1-1 provides an opportunity to further dissect the role of SMA1 in miRNA biogenesis, splicing and transcription in the near future. The discovery that SMA1 functions in miRNA biogenesis and transcription has expanded the knowledge on the function of Prp28. It will be worth to test if Prp28 has the similar roles in other organisms.
DATA AVAILABILITY
Multiple sequence alignment was performed with Clustal X (75) (http://www.clustal.org). Phylogenetic tree was constructed using MEGA 7 (76) (http://www.megasoftware.net). The statistically significant differences were analyzed with STATA software (https://www.stata.com).
Supplementary Material
ACKNOWLEDGEMENTS
We acknowledge the NASC for providing the sma1-2 (SALK_110742) seeds.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Nebraska Soybean Board [1727 to B.Y.]; National Science Foundation [MCB1808182 to B.Y.]; National Natural Science Foundation of China [31425004, 91417304, 91017014 to Y.L.; 31400249 to R.X.]; Ministry of Agriculture of China [2016ZX08009-003 to N.L.]; Strategic Priority Research Program ‘Molecular Mechanism of Plant Growth and Development’ [XDBP401 to Y.L.]; Pioneer Hundred Program of Chinese Academy of Sciences (to S.L.). Funding for open access charge: Pioneer Hundred Program of Chinese Academy of Sciences.
Conflict of interest statement. None declared.
REFERENCES
- 1. Chen X. Small RNAs and their roles in plant development. Annu. Rev. Cell Dev. Biol. 2009; 25:21–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Axtell M.J., Snyder J.A., Bartel D.P.. Common functions for diverse small RNAs of land plants. Plant Cell. 2007; 19:1750–1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004; 116:281–297. [DOI] [PubMed] [Google Scholar]
- 4. Li S., Castillo-Gonzalez C., Yu B., Zhang X.. The functions of plant small RNAs in development and in stress responses. Plant J. 2017; 90:654–670. [DOI] [PubMed] [Google Scholar]
- 5. Xie Z., Allen E., Fahlgren N., Calamar A., Givan S.A., Carrington J.C.. Expression of Arabidopsis MIRNA genes. Plant Physiol. 2005; 138:2145–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Baulcombe D. RNA silencing in plants. Nature. 2004; 431:356–363. [DOI] [PubMed] [Google Scholar]
- 7. Kurihara Y., Watanabe Y.. Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. PNAS. 2004; 101:12753–12758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Qi Y., Denli A.M., Hannon G.J.. Biochemical specialization within Arabidopsis RNA silencing pathways. Mol. Cell. 2005; 19:421–428. [DOI] [PubMed] [Google Scholar]
- 9. Dong Z., Han M.H., Fedoroff N.. The RNA-binding proteins HYL1 and SE promote accurate in vitro processing of pri-miRNA by DCL1. PNAS. 2008; 105:9970–9975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lobbes D., Rallapalli G., Schmidt D.D., Martin C., Clarke J.. SERRATE: a new player on the plant microRNA scene. EMBO Rep. 2006; 7:1052–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kurihara Y., Takashi Y., Watanabe Y.. The interaction between DCL1 and HYL1 is important for efficient and precise processing of pri-miRNA in plant microRNA biogenesis. RNA. 2006; 12:206–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yu B., Yang Z., Li J., Minakhina S., Yang M., Padgett R.W., Steward R., Chen X.. Methylation as a crucial step in plant microRNA biogenesis. Science. 2005; 307:932–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhai J., Zhao Y., Simon S.A., Huang S., Petsch K., Arikit S., Pillay M., Ji L., Xie M., Cao X. et al. . Plant microRNAs display differential 3′ truncation and tailing modifications that are ARGONAUTE1 dependent and conserved across species. Plant Cell. 2013; 25:2417–2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ren G., Xie M., Zhang S., Vinovskis C., Chen X., Yu B.. Methylation protects microRNAs from an AGO1-associated activity that uridylates 5′ RNA fragments generated by AGO1 cleavage. PNAS. 2014; 111:6365–6370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Park M.Y., Wu G., Gonzalez-Sulser A., Vaucheret H., Poethig R.S.. Nuclear processing and export of microRNAs in Arabidopsis. PNAS. 2005; 102:3691–3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Earley K.W., Poethig R.S.. Binding of the cyclophilin 40 ortholog SQUINT to Hsp90 protein is required for SQUINT function in Arabidopsis. J. Biol. Chem. 2011; 286:38184–38189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cui Y., Fang X., Qi Y.. TRANSPORTIN1 Promotes the Association of MicroRNA with ARGONAUTE1 in Arabidopsis. Plant Cell. 2016; 28:2576–2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mi S., Cai T., Hu Y., Chen Y., Hodges E., Ni F., Wu L., Li S., Zhou H., Long C. et al. . Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell. 2008; 133:116–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kim Y.J., Zheng B., Yu Y., Won S.Y., Mo B., Chen X.. The role of Mediator in small and long noncoding RNA production in Arabidopsis thaliana. EMBO J. 2011; 30:814–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang L., Song X., Gu L., Li X., Cao S., Chu C., Cui X., Chen X., Cao X.. NOT2 proteins promote polymerase II-dependent transcription and interact with multiple MicroRNA biogenesis factors in Arabidopsis. Plant Cell. 2013; 25:715–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang S., Xie M., Ren G., Yu B.. CDC5, a DNA binding protein, positively regulates posttranscriptional processing and/or transcription of primary microRNA transcripts. PNAS. 2013; 110:17588–17593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fang X., Cui Y., Li Y., Qi Y.. Transcription and processing of primary microRNAs are coupled by Elongator complex in Arabidopsis. Nat. Plants. 2015; 1:15075. [DOI] [PubMed] [Google Scholar]
- 23. Hajheidari M., Farrona S., Huettel B., Koncz Z., Koncz C.. CDKF;1 and CDKD protein kinases regulate phosphorylation of serine residues in the C-terminal domain of Arabidopsis RNA polymerase II. Plant Cell. 2012; 24:1626–1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yu B., Bi L., Zheng B., Ji L., Chevalier D., Agarwal M., Ramachandran V., Li W., Lagrange T., Walker J.C. et al. . The FHA domain proteins DAWDLE in Arabidopsis and SNIP1 in humans act in small RNA biogenesis. PNAS. 2008; 105:10073–10078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhang S., Liu Y., Yu B.. PRL1, an RNA-binding protein, positively regulates the accumulation of miRNAs and siRNAs in Arabidopsis. PLos Genet. 2014; 10:e1004841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ren G., Xie M., Dou Y., Zhang S., Zhang C., Yu B.. Regulation of miRNA abundance by RNA binding protein TOUGH in Arabidopsis. PNAS. 2012; 109:12817–12821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Francisco-Mangilet A.G., Karlsson P., Kim M.H., Eo H.J., Oh S.A., Kim J.H., Kulcheski F.R., Park S.K., Manavella P.A.. THO2, a core member of the THO/TREX complex, is required for microRNA production in Arabidopsis. Plant J. 2015; 82:1018–1029. [DOI] [PubMed] [Google Scholar]
- 28. Li S., Liu K., Zhang S., Wang X., Rogers K., Ren G., Zhang C., Yu B.. STV1, a ribosomal protein, binds primary microRNA transcripts to promote their interaction with the processing complex in Arabidopsis. PNAS. 2017; 114:1424–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jia T., Zhang B., You C., Zhang Y., Zeng L., Li S., Johnson K.C.M., Yu B., Li X., Chen X.. The Arabidopsis MOS4-associated complex promotes microRNA biogenesis and precursor messenger RNA Splicing. Plant Cell. 2017; 29:2626–2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Manavella P.A., Hagmann J., Ott F., Laubinger S., Franz M., Macek B., Weigel D.. Fast-forward genetics identifies plant CPL phosphatases as regulators of miRNA processing factor HYL1. Cell. 2012; 151:859–870. [DOI] [PubMed] [Google Scholar]
- 31. Yan J., Wang P., Wang B., Hsu C.C., Tang K., Zhang H., Hou Y.J., Zhao Y., Wang Q., Zhao C. et al. . The SnRK2 kinases modulate miRNA accumulation in Arabidopsis. PLos Genet. 2017; 13:e1006753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cho S.K., Ben Chaabane S., Shah P., Poulsen C.P., Yang S.W.. COP1 E3 ligase protects HYL1 to retain microRNA biogenesis. Nat. Commun. 2014; 5:5867. [DOI] [PubMed] [Google Scholar]
- 33. Zhang Z., Guo X., Ge C., Ma Z., Jiang M., Li T., Koiwa H., Yang S.W., Zhang X.. KETCH1 imports HYL1 to nucleus for miRNA biogenesis in Arabidopsis. PNAS. 2017; 114:4011–4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ben Chaabane S., Liu R., Chinnusamy V., Kwon Y., Park J.H., Kim S.Y., Zhu J.K., Yang S.W., Lee B.H.. STA1, an Arabidopsis pre-mRNA processing factor 6 homolog, is a new player involved in miRNA biogenesis. Nucleic Acids Res. 2013; 41:1984–1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fang X., Shi Y., Lu X., Chen Z., Qi Y.. CMA33/XCT regulates small RNA production through modulating the transcription of dicer-like genes in Arabidopsis. Mol. Plant. 2015; 8:1227–1236. [DOI] [PubMed] [Google Scholar]
- 36. Gregory B.D., O’Malley R.C., Lister R., Urich M.A., Tonti-Filippini J., Chen H., Millar A.H., Ecker J.R.. A link between RNA metabolism and silencing affecting Arabidopsis development. Dev. Cell. 2008; 14:854–866. [DOI] [PubMed] [Google Scholar]
- 37. Laubinger S., Sachsenberg T., Zeller G., Busch W., Lohmann J.U., Ratsch G., Weigel D.. Dual roles of the nuclear cap-binding complex and SERRATE in pre-mRNA splicing and microRNA processing in Arabidopsis thaliana. PNAS. 2008; 105:8795–8800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mathew R., Hartmuth K., Mohlmann S., Urlaub H., Ficner R., Luhrmann R.. Phosphorylation of human PRP28 by SRPK2 is required for integration of the U4/U6-U5 tri-snRNP into the spliceosome. Nat. Struct. Mol. Biol. 2008; 15:435–443. [DOI] [PubMed] [Google Scholar]
- 39. Jacewicz A., Schwer B., Smith P., Shuman S.. Crystal structure, mutational analysis and RNA-dependent ATPase activity of the yeast DEAD-box pre-mRNA splicing factor Prp28. Nucleic Acids Res. 2014; 42:12885–12898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Liu Y.C., Cheng S.C.. Functional roles of DExD/H-box RNA helicases in pre-mRNA splicing. J. Biomed. Sci. 2015; 22:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Strauss E.J., Guthrie C.. PRP28, a ‘DEAD-box’ protein, is required for the first step of mRNA splicing in vitro. Nucleic Acids Res. 1994; 22:3187–3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Konishi T., Uodome N., Sugimoto A.. The Caenorhabditis elegans DDX-23, a homolog of yeast splicing factor PRP28, is required for the sperm-oocyte switch and differentiation of various cell types. Dev. Dyn. 2008; 237:2367–2377. [DOI] [PubMed] [Google Scholar]
- 43. Wang Z.P., Xing H.L., Dong L., Zhang H.Y., Han C.Y., Wang X.C., Chen Q.J.. Egg cell-specific promoter-controlled CRISPR/Cas9 efficiently generates homozygous mutants for multiple target genes in Arabidopsis in a single generation. Genome Biol. 2015; 16:144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Li S., Liu Y., Zheng L., Chen L., Li N., Corke F., Lu Y., Fu X., Zhu Z., Bevan M.W. et al. . The plant-specific G protein gamma subunit AGG3 influences organ size and shape in Arabidopsis thaliana. New Phytol. 2012; 194:690–703. [DOI] [PubMed] [Google Scholar]
- 45. Mohlmann S., Mathew R., Neumann P., Schmitt A., Luhrmann R., Ficner R.. Structural and functional analysis of the human spliceosomal DEAD-box helicase Prp28. Acta Crystallogr. D. Biol. Crystallogr. 2014; 70:1622–1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Phatnani H.P., Greenleaf A.L.. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 2006; 20:2922–2936. [DOI] [PubMed] [Google Scholar]
- 47. Hajheidari M., Koncz C., Eick D.. Emerging roles for RNA polymerase II CTD in Arabidopsis. Trends Plant Sci. 2013; 18:633–643. [DOI] [PubMed] [Google Scholar]
- 48. Bowman E.A., Kelly W.G.. RNA polymerase II transcription elongation and Pol II CTD Ser2 phosphorylation: a tail of two kinases. Nucleus. 2014; 5:224–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Li S., Liu K., Zhou B., Li M., Zhang S., Zeng L., Zhang C., Yu B.. MAC3A and MAC3B, two core subunits of the MOS4-associated complex, positively influence miRNA biogenesis. Plant Cell. 2018; 30:481–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wu X., Shi Y., Li J., Xu L., Fang Y., Li X., Qi Y.. A role for the RNA-binding protein MOS2 in microRNA maturation in Arabidopsis. Cell Res. 2013; 23:645–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Li S., Liu K., Zhou B., Li M., Zhang S., Zeng L., Zhang C., Yu B.. MAC3A and MAC3B, two core subunits of the MOS4-associated complex, positively influence miRNA Biogenesis. Plant Cell. 2018; 30:481–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Strauss E.J., Guthrie C.. A cold-sensitive mRNA splicing mutant is a member of the RNA helicase gene family. Genes Dev. 1991; 5:629–641. [DOI] [PubMed] [Google Scholar]
- 53. Teigelkamp S., Mundt C., Achsel T., Will C.L., Luhrmann R.. The human U5 snRNP-specific 100-kD protein is an RS domain-containing, putative RNA helicase with significant homology to the yeast splicing factor Prp28p. RNA. 1997; 3:1313–1326. [PMC free article] [PubMed] [Google Scholar]
- 54. Chen J.Y., Stands L., Staley J.P., Jackups R.R. Jr, Latus L.J., Chang T.H.. Specific alterations of U1-C protein or U1 small nuclear RNA can eliminate the requirement of Prp28p, an essential DEAD box splicing factor. Mol. Cell. 2001; 7:227–232. [DOI] [PubMed] [Google Scholar]
- 55. Staley J.P., Guthrie C.. An RNA switch at the 5′ splice site requires ATP and the DEAD box protein Prp28p. Mol. Cell. 1999; 3:55–64. [DOI] [PubMed] [Google Scholar]
- 56. Mattioli C., Pianigiani G., Pagani F.. Cross talk between spliceosome and microprocessor defines the fate of pre-mRNA. Wiley Interdiscip. Rev. RNA. 2014; 5:647–658. [DOI] [PubMed] [Google Scholar]
- 57. Stepien A., Knop K., Dolata J., Taube M., Bajczyk M., Barciszewska-Pacak M., Pacak A., Jarmolowski A., Szweykowska-Kulinska Z.. Posttranscriptional coordination of splicing and miRNA biogenesis in plants. Wiley Interdiscip. Rev. RNA. 2017; 8:e1403. [DOI] [PubMed] [Google Scholar]
- 58. Raczynska K.D., Stepien A., Kierzkowski D., Kalak M., Bajczyk M., McNicol J., Simpson C.G., Szweykowska-Kulinska Z., Brown J.W., Jarmolowski A.. The SERRATE protein is involved in alternative splicing in Arabidopsis thaliana. Nucleic Acids Res. 2014; 42:1224–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Chen T., Cui P., Chen H., Ali S., Zhang S., Xiong L.. A KH-domain RNA-binding protein interacts with FIERY2/CTD phosphatase-like 1 and splicing factors and is important for pre-mRNA splicing in Arabidopsis. PLos Genet. 2013; 9:e1003875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Koster T., Meyer K., Weinholdt C., Smith L.M., Lummer M., Speth C., Grosse I., Weigel D., Staiger D.. Regulation of pri-miRNA processing by the hnRNP-like protein AtGRP7 in Arabidopsis. Nucleic Acids Res. 2014; 42:9925–9936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Karlsson P., Christie M.D., Seymour D.K., Wang H., Wang X., Hagmann J., Kulcheski F., Manavella P.A.. KH domain protein RCF3 is a tissue-biased regulator of the plant miRNA biogenesis cofactor HYL1. PNAS. 2015; 112:14096–14101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Hirsch J., Lefort V., Vankersschaver M., Boualem A., Lucas A., Thermes C., d’Aubenton-Carafa Y., Crespi M.. Characterization of 43 non-protein-coding mRNA genes in Arabidopsis, including the MIR162a-derived transcripts. Plant Physiol. 2006; 140:1192–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Brown J.W., Marshall D.F., Echeverria M.. Intronic noncoding RNAs and splicing. Trends Plant Sci. 2008; 13:335–342. [DOI] [PubMed] [Google Scholar]
- 64. Lu C., Jeong D.H., Kulkarni K., Pillay M., Nobuta K., German R., Thatcher S.R., Maher C., Zhang L., Ware D. et al. . Genome-wide analysis for discovery of rice microRNAs reveals natural antisense microRNAs (nat-miRNAs). PNAS. 2008; 105:4951–4956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Yan K., Liu P., Wu C.A., Yang G.D., Xu R., Guo Q.H., Huang J.G., Zheng C.C.. Stress-induced alternative splicing provides a mechanism for the regulation of microRNA processing in Arabidopsis thaliana. Mol. Cell. 2012; 48:521–531. [DOI] [PubMed] [Google Scholar]
- 66. Bielewicz D., Kalak M., Kalyna M., Windels D., Barta A., Vazquez F., Szweykowska-Kulinska Z., Jarmolowski A.. Introns of plant pri-miRNAs enhance miRNA biogenesis. EMBO Rep. 2013; 14:622–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Schwab R., Speth C., Laubinger S., Voinnet O.. Enhanced microRNA accumulation through stemloop-adjacent introns. EMBO Rep. 2013; 14:615–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Jia F., Rock C.D.. MIR846 and MIR842 comprise a cistronic MIRNA pair that is regulated by abscisic acid by alternative splicing in roots of Arabidopsis. Plant Mol. Biol. 2013; 81:447–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Zielezinski A., Dolata J., Alaba S., Kruszka K., Pacak A., Swida-Barteczka A., Knop K., Stepien A., Bielewicz D., Pietrykowska H. et al. . mirEX 2.0—an integrated environment for expression profiling of plant microRNAs. BMC Plant Biol. 2015; 15:144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Szarzynska B., Sobkowiak L., Pant B.D., Balazadeh S., Scheible W.R., Mueller-Roeber B., Jarmolowski A., Szweykowska-Kulinska Z.. Gene structures and processing of Arabidopsis thaliana HYL1-dependent pri-miRNAs. Nucleic Acids Res. 2009; 37:3083–3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Knop K., Stepien A., Barciszewska-Pacak M., Taube M., Bielewicz D., Michalak M., Borst J.W., Jarmolowski A., Szweykowska-Kulinska Z.. Active 5′ splice sites regulate the biogenesis efficiency of Arabidopsis microRNAs derived from intron-containing genes. Nucleic Acids Res. 2017; 45:2757–2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Hong S., Noh H., Chen H., Padia R., Pan Z.K., Su S.B., Jing Q., Ding H.F., Huang S.. Signaling by p38 MAPK stimulates nuclear localization of the microprocessor component p68 for processing of selected primary microRNAs. Sci. Signal. 2013; 6:ra16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Remenyi J., Bajan S., Fuller-Pace F.V., Arthur J.S., Hutvagner G.. The loop structure and the RNA helicase p72/DDX17 influence the processing efficiency of the mice miR-132. Sci. Rep. 2016; 6:22848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Yin J., Park G., Lee J.E., Choi E.Y., Park J.Y., Kim T.H., Park N., Jin X., Jung J.E., Shin D. et al. . DEAD-box RNA helicase DDX23 modulates glioma malignancy via elevating miR-21 biogenesis. Brain. 2015; 138:2553–2570. [DOI] [PubMed] [Google Scholar]
- 75. Chenna R., Sugawara H., Koike T., Lopez R., Gibson T.J., Higgins D.G., Thompson J.D.. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003; 31:3497–3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Kumar S., Stecher G., Tamura K.. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016; 33:1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Multiple sequence alignment was performed with Clustal X (75) (http://www.clustal.org). Phylogenetic tree was constructed using MEGA 7 (76) (http://www.megasoftware.net). The statistically significant differences were analyzed with STATA software (https://www.stata.com).