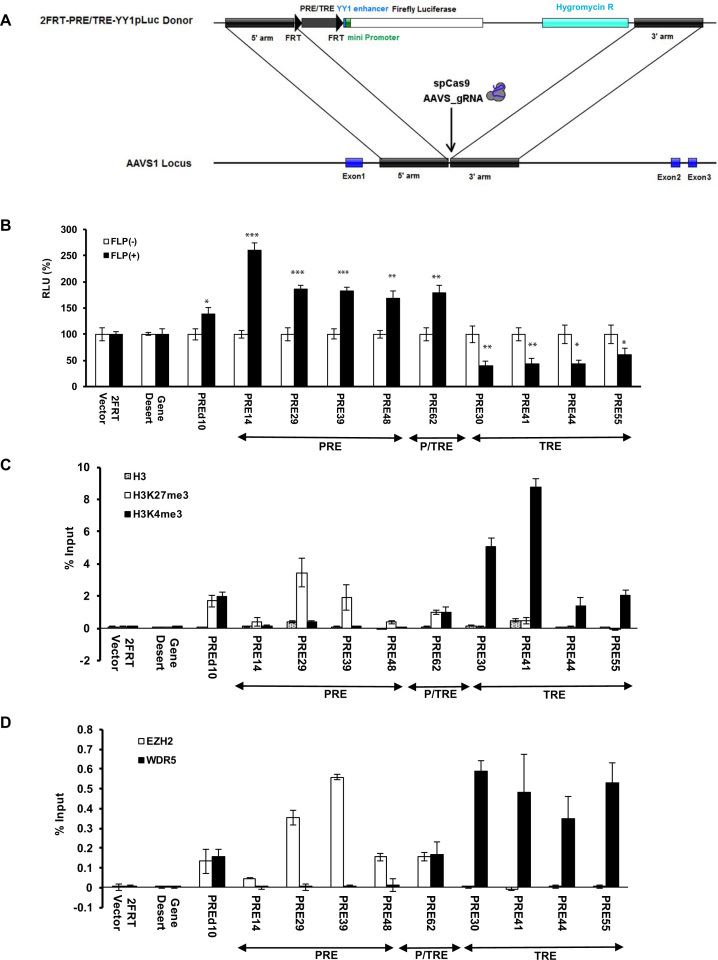
Figure 5.
Effects of putative PREs and TREs on transcription in an identical genomic environment. A). Schematic illustration of the strategy that integrated the core sequences of the putative PREs and TREs (2FRT-PRE/TRE-YY1pLuc donor) into AAVS1 locus by CRISPR-Cas9. B). Dual luciferases assay in HeLa cells carrying stably integrated core sequences comparing FLP(-) samples (open bars) and FLP(+) samples (filled solid bars). Data are represented as mean ± SD from at least three replicates. The P value of Student's t-test comparing FLP(+) and FLP(-) for each integrated core sequence was shown as * for P<0.05, ** for P<0.005 and *** for P<0.001. C). ChIP-qPCR of H3 (dotted open bars), H3K27me3 (open bars) and H3K4me3 (filled solid bars) at the promoter region downstream of the integrated core sequences shown as % input. D). ChIP-qPCR results of EZH2 (open bars) and WDR5 (filled solid bars) at the promoter region downstream of the integrated core sequences shown as % input.