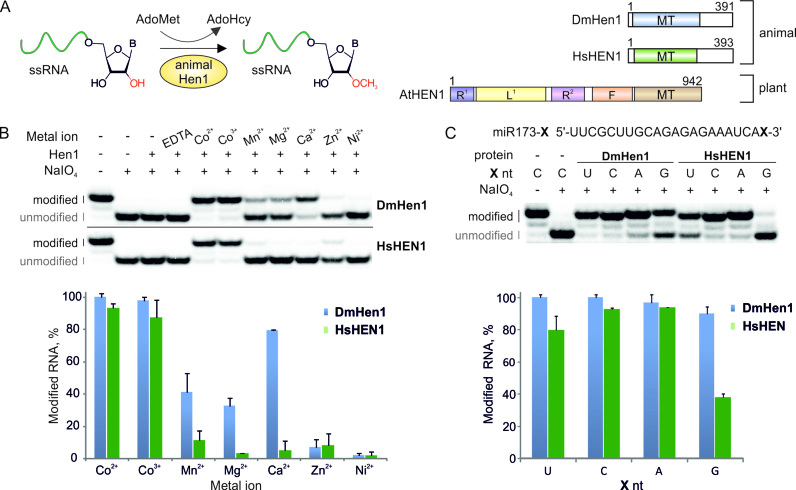
Figure 1.
DmHen1 and HsHEN1 methyltransferases require cobalt cations for efficient modification of terminal nucleosides of single-stranded RNA. (A) Schematic representation of animal Hen1 methylation reaction together with structures of Drosophila melanogaster DmHen1, Homo sapiens HsHEN1 and Arabidopsis thaliana AtHEN1 methyltransferases. MT: methyltransferase domain; R1 and R2: dsRNA binding domains; L: La-motif-containing domain; F: FK506 binding protein-like domain. (B) Only the presence of Co2+ or Co3+ confers full methylation of RNA substrate. Reactions were performed using 0.2 μM of 5′-32P-labelled 22-nt miR173, 0.1 mM of AdoMet 1, 10 mM of metal ion in the form of chloride salt (CoCl2 or [Co(NH2)6]Cl3 in the case of Co2+ or Co3+, respectively), except for nickel sulphate and 1 μM of Hen1 proteins. After 30 min of incubation at 37°C, the extent of modification was determined by NaIO4-mediated oxidation/β-elimination as described in (24). This treatment resulted in shortening of unmodified RNA by 1 nt and increasing its mobility relatively to the modified one. Modified and unmodified RNA strands are marked in black and grey, accordingly. Histogram of RNA methylation efficiency with different metal cofactors was calculated from duplicate experiments. (C) Co2+ reduces 3′ terminal nucleoside bias in DmHen1 methylation reactions. miR173 with different 3′ terminal nucleosides, namely U, C, A and G, was used as a substrate for DmHen1 or HsHEN1. Results presented in a histogram are average of two experiments.