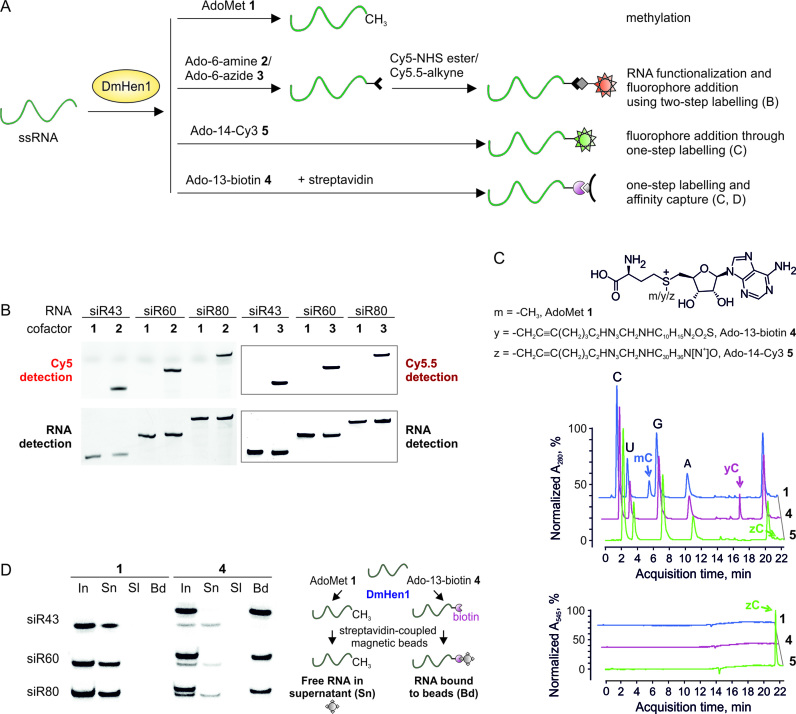
Figure 5.
DmHen1-directed functionalization of various length single-stranded RNAs. (A) Principal scheme of two- and one-step ssRNA labelling in comparison to natural reaction of DmHen1. (B) Visualization of ssRNAs labelled by two-step approach on a denaturing PAA gel. 0.2 μM of ssRNAs incubated with 1 μM of DmHen1 and 0.1 mM of Ado-6-amine 2 or Ado-6-azide 3, were treated with Cy5-649/670-NHS ester or Cy5.5-673/707-alkyne, respectively. RNAs methylated using AdoMet 1 served as a control for specific labelling. Cy5 and Cy5.5 fluorescence was detected using 635 and 670 nm lasers accordingly (top panel), bulk RNA was visualized after staining with ethidium bromide (bottom panel). The detailed scheme of current experiment can be found in Supplementary Figure S6. (C) RP-HPLC analysis of nucleosides derived after one-step labelling of miR173 by DmHen1. The top chromatogram shows the absorption at 280 nm, whereas the bottom one highlights the absorption of C attached Cy3 fluorophore at 545 nm. 1 μM of methyltransferase was used to modify 2 μM of miR173 in the presence of 100 μM of AdoMet 1, 50 μM of Ado-13-biotin 4 or 6 μM of Ado-14-Cy3 5. After 1.5 h of incubation at 37°C RNA was degraded to nucleotides, dephosphorylated and analysed by RP-HPLC. (D) Affinity capture of ssRNAs following one-step labelling. 50 μM of Ado-13-biotin 4 was used to label 0.01 μM of RNA substrate in the presence of 1 μM DmHen1. The initial mixture of biotin labelled RNAs (In) was loaded on streptavidin beads and supernatant (Sn) was collected. Following a buffer wash (Sl), streptavidin beads were resuspended in 95% formamide, 10 mM EDTA and heated for 10 min at 70°C to release bound RNA (Bd). As a control ssRNAs were modifies with 100 μM AdoMet 1. The right panel represents an experimental outline.