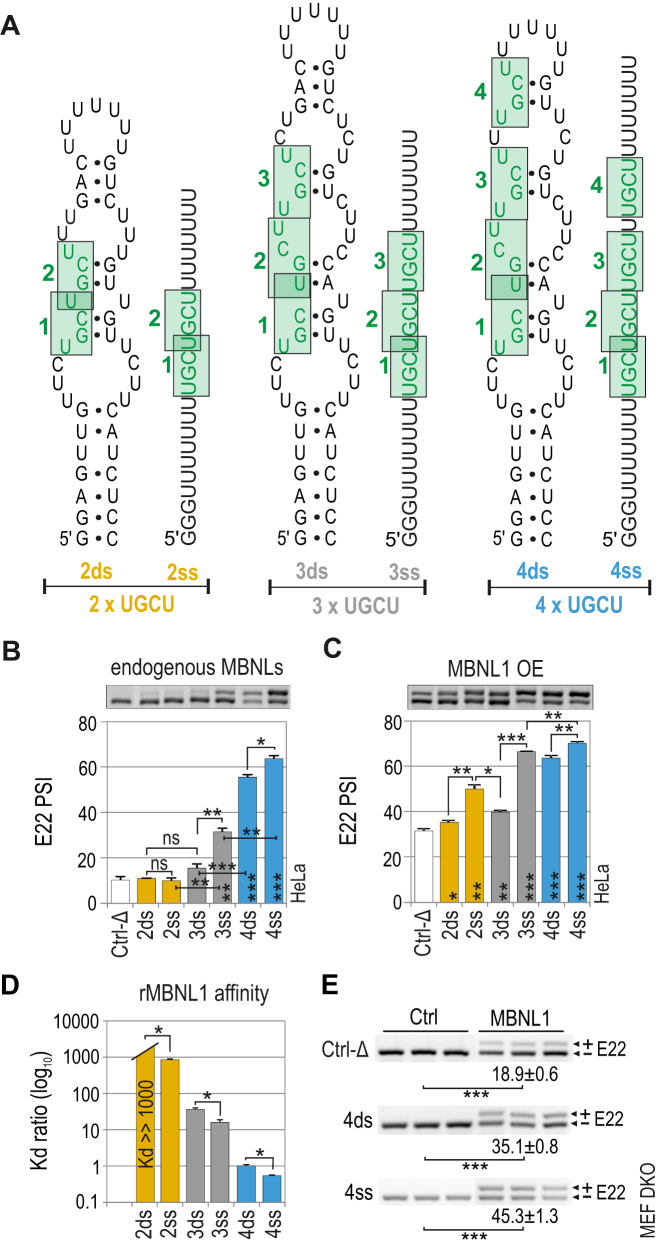
Figure 2.
MBNL1 binding and splicing activity is regulated by both the number of UGCU motifs and structural context. (A) RNA secondary structures of semi-stable (ds; double-stranded) and unstructured (ss; single-stranded) artificial regulatory elements which are base paired at the 5′ and 3′ ends and internally (only ds RNA) to force the formation of one thermodynamically optimal secondary structure confirmed by us in several RNA structure prediction software programs (46–48). MBNL-binding motifs are numbered from 5′-end and marked in green. The RNA names correspond to the number of UGCU motifs (2, 3 or 4) and the structure type (ds or ss). The splicing response of pre-mRNAs of hybrid minigenes with ds or ss RNAs incorporated in I22 upon (B) endogenous level of MBNLs or (C) MBNL1 overexpression (OE) in HeLa cells. Vertical asterisks denote the statistical significance of results in comparison to a control experiment (Ctrl-Δ; transfection with Atp1a1-Δ minigene). Note a positive effect of the increasing UGCU motif number on alternative splicing - the PSI of E22 for 2ss < 3ss < 4ss; MBNL1 dose-dependency of weaker regulatory elements - the PSI of E22 for 2ds < 2ss > 3ds; and the structural organization of an RNA regulatory element surpassing the number of UGCU motifs - PSI of E22 for 4ds = 3ss; 2ss > 3ds; n = 2. (D) Quantification of the biochemical assay showing relative rMBNL1 binding affinity for RNA fragments normalized to Kd for the 4ds RNA molecule; n = 2 for each protein concentration (in the range of 0–200 nM of rMBNL1). (E) RT-PCR showing the splicing response of hybrid Atp2a1 minigenes representing ss and ds groups of RNAs with or without MBNL1 OE in Mbnl1; Mbnl2 DKO MEFs. The asterisks denote the statistical significance of results compared to cells treated with control eGFP construct (Ctrl). Ctrl-Δ, transfection with Atp1a1-Δ minigene; n = 3.