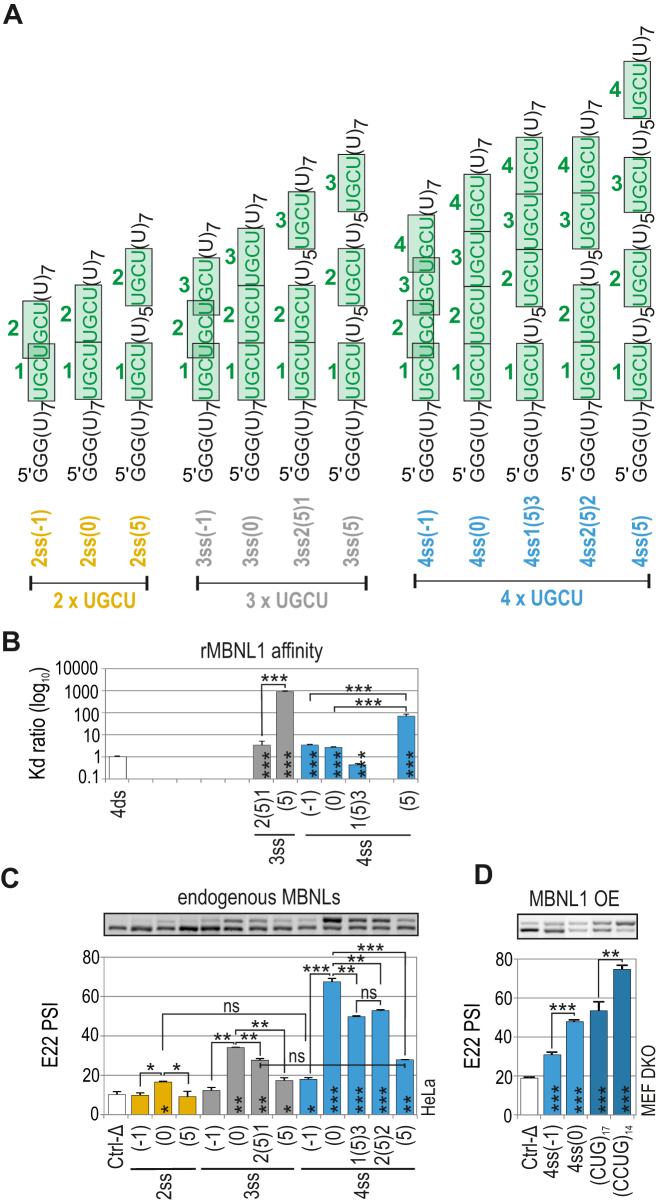
Figure 3.
Distance between UGCU motifs modulates MBNL-mediated splicing regulation but not binding affinity. (A) The structures of ss RNA regulatory elements with distinctly interspaced UGCU motifs marked in green. The RNA names correspond to the distance between consecutive UGCU motifs, which is marked in brackets surrounded by the number of adjacent motifs. (B) As in Figure 2D but for a group of ss RNAs with interspaced UGCU motifs; n = 2–4 for each protein concentration (in the range of 0–400 nM of rMBNL1). (C) As in Figure 2C but for a group of ss RNAs with interspaced UGCU motifs; n = 2. Note no or very weak regulatory properties of RNAs with overlapping motifs - 2ss(–1), 3ss(–1) and 4ss(–1). RNAs with sequential motifs - 4ss(0), 3ss(0) and 2ss(0), exceeded all other tested regulatory elements within the UGCU number sets. UGCU separation had an adverse effect on E22 inclusion - the PSI of E22 for 4ss(0) > 4ss3(5)1 = 4ss2(5)2 > 4ss(5). Note the redundant impact of UGCU number - PSI of E22 for 3ss2(5)1 = 4ss(5). Vertical asterisks denote the statistical significance of results in comparison to a control experiment (Ctrl-Δ; transfection with Atp1a1-Δ minigene). (D) The splicing response of hybrid Atp2a1 minigenes with regulatory elements distinguished by either overlapping or sequential arrangement of distinct number of UGCU motifs. The graph shows RT-PCR results for DKO MEFs co-transfected with hybrid minigenes and the MBNL1 expression vector. Vertical asterisks denote the statistical significance of results in comparison to Ctrl-Δ construct; n = 3.