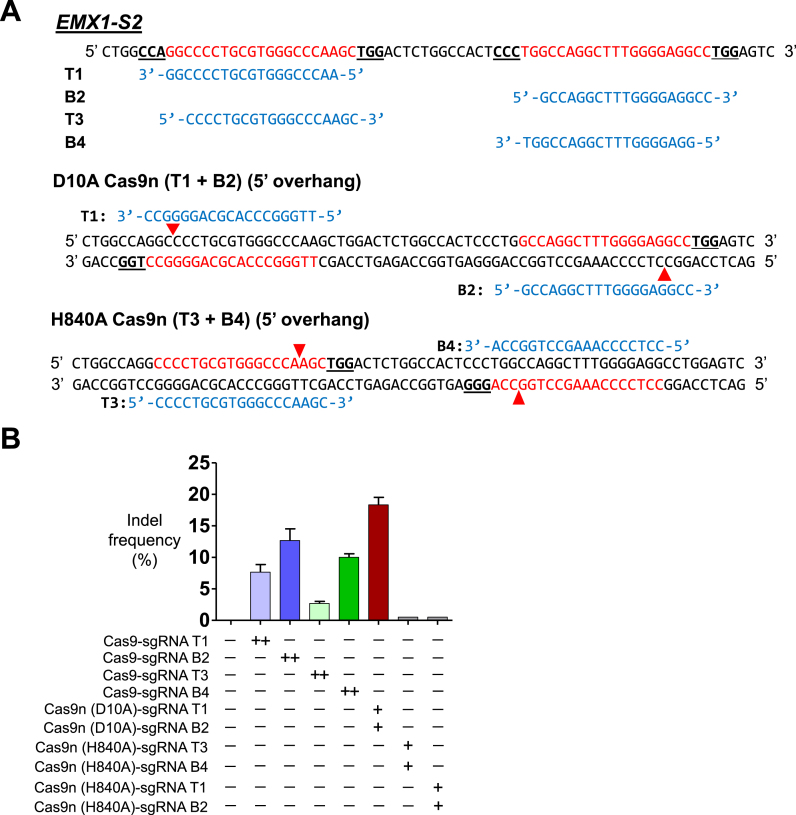
Figure 5.
Comparison of indel generation efficiencies of paired Cas9 D10A nickases versus paired Cas9 H840A nickases using reverse-identical 5′GX19 sgRNAs without control sgRNA. (A) Sequence of the human EMX1-S2 locus with designed sgRNAs (T1, B2, T3 and B4). Reverse-identical sgRNAs (T1 and T3; B2 and B4) are shown in blue letters. sgRNA target sites (T1, B2, T3 and B4) are indicated by red letters and PAM sequences are marked by bold underlined letters. The cleavage by D10A Cas9 nickase and the sgRNA pair of T1 and B2 leads to a double-strand break with a 5′ overhang. Similarly, the cleavage by H840A Cas9 nickase and the sgRNA pair of T3 and B4 also leads to a double strand break with a 5′ overhang. Red arrowheads indicate the cleavage site. (B) The frequency of double the amount of Cas9 nuclease- or paired nickase-driven mutations as determined by the T7E1 assay are shown using bar graph. Error bars were derived from three independent experiments (n = 3). ‘+’ and ‘++’ denote 1 and 2 μg concentrations of Cas9 nucleases or paired Cas9 nickases using top or bottom sgRNAs, respectively.