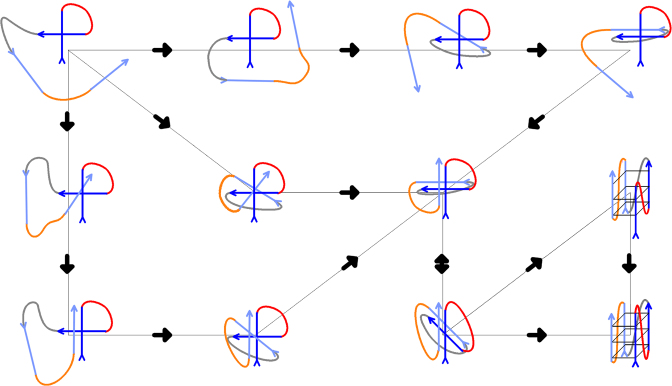
Figure 8.
Sketch of a possible folding pathway for intramolecular three-quartet RNA GQ starting from unstructured single strand (not shown in the scheme); for more detailed view on folding pathway of PHs see Figure 7. Directionality of the chain is marked by arrows. G-strands are in blue (first and second in dark blue, third and fourth in light blue) while loops one, two and three are in red, grey and orange, respectively. The essence of the process is formation of compact coil-like structures stabilized by extensive cross-like interactions among the four G-strands. The PHs and propeller loops can form via conformational diffusion (see the text). Conformational diffusion starting from the coil-like structures may allow structuring of slipped GQs with reduced number of quartets (see refs. (66) and (35) for more information) or direct formation of the native parallel GQ. Note that the scheme is inevitably oversimplified and aims to illustrate the suggested principle of the folding rather than any exact pathway. The Figure nevertheless reflects all the knowledge that has been gathered so far by atomistic MD about properties of various types of structures that contribute to the GQ folding landscapes. The scheme shows a variant of folding that proceeds via an initial cross-like interaction between the first two strands. The Figure does not depict incorporation of the monovalent ions, however, involvement of the ions in the process is suggested to take place once extensive GQ-like GG H-bonding patterns are formed. In any case, ions should be specifically bound once first complete G-quartet forms. The scheme does not depict participation of conventional GQ-like parallel triplexes (38,109,110), as these are assumed to be more transient than the structures included in the Figure (24,38). Note that while the MD simulation method allows to suggest plausible types of structures that may be present on the folding landscape, it is not yet possible to simulate the full folding process (24). Therefore, we do not provide any estimate of the timescale of the folding (and of the relative contribution of coil-like structures and strand-slipped GQs) and its determination awaits appropriate experimental measurements. The rearrangements depicted should exist also on the free-energy (folding) landscapes of DNA GQs, which are nevertheless much more complex due to presence of deep competing basins created by alternative GQ topologies and dominating kinetic partitioning (24).