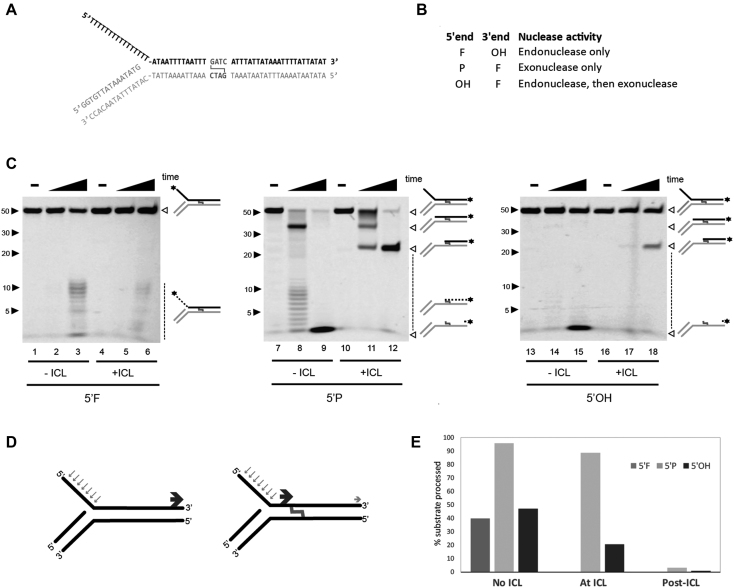
Figure 4.
SNM1A can generate a nick to initiate trimming to the crosslink. (A) DNA substrate design mimicking stalled replication fork. (B) Substrate end modifications and corresponding nuclease activities monitored. F, fluorophore; OH, hydroxyl; P, phosphate. (C) In vitro analysis of SNM1A nuclease activities on 5′flap crosslinked DNA. SNM1A (0.4 μM) was incubated with substrate (0.1μM) as shown in (A) with or without an SJG-136 crosslink. Substrates are labeled as indicated in (B). Reactions were stopped after 0, 2 and 30 min. Products were resolved using 20% denaturing PAGE and imaged with the ChemiDoc XRS (BioRad) at 526 nm. (D) Summary of nuclease events on 5′ flap substrate with or without an SJG crosslink. Blue arrows indicate initial endonuclease cleavage. Dark gray arrows indicate exonuclease product. Light grey arrow indicates translesional nuclease product. (E) Quantification of product formed in the absence of the ICL, at the ICL, or past the ICL. Products were quantified using ImageLab.