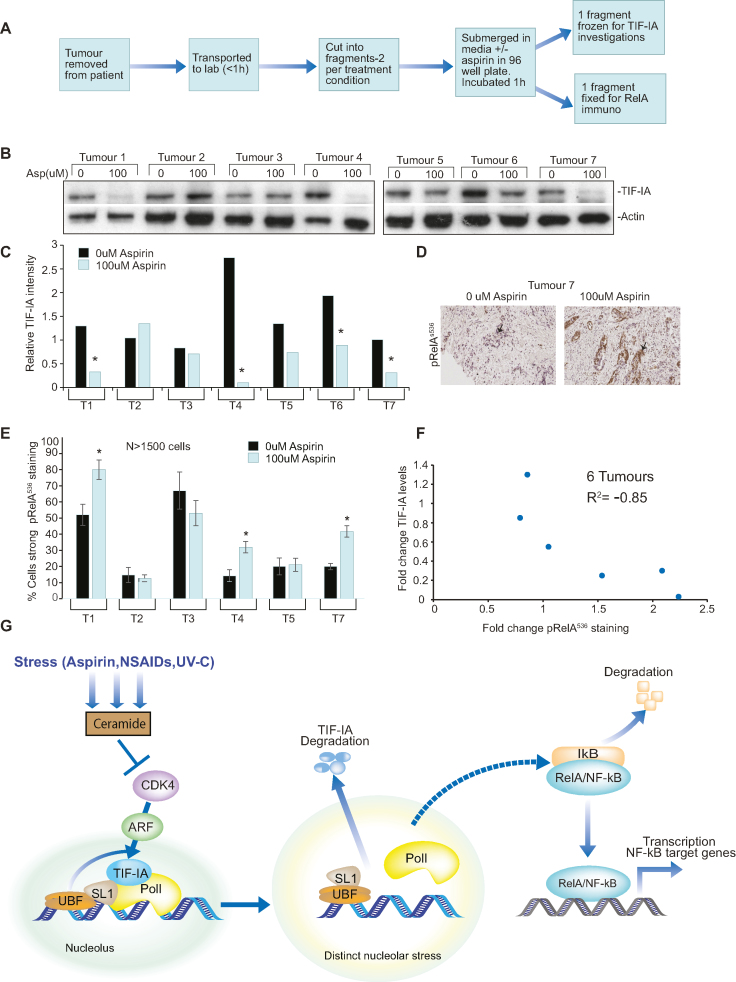
Figure 9.
TIF-IA degradation correlates with NF-κB pathway activation in human clinical samples. (A) Diagram depicting workflow of ex vivo culture. Resected colorectal tumour biopsies were immediately transferred to the lab, washed, immersed in culturing media in 96-well plates then exposed to 0 or 100 uM aspirin for 1 h. One piece of tissue was fixed for immunohistochemistry while another was frozen for protein analysis. This was carried out for seven patients. (B) Immunoblot was performed on WCL with the indicated antibodies. (C) ImageJ quantified TIF-IA intensity relative to actin. * Tumours showing a >2-fold decrease in relative levels of TIF-IA in response to aspirin were deemed to respond. (D and E) Immunohistochemistry was performed on sections from paraffin embedded tissue with antibodies to RelAp536. (D) An example immunomicrograph. Arrows indicate epithelial cells. (E) Leica QWin plus image analysis software, with scripts written in house, was used to quantify the nuclear RelAp536 intensity in digitized images. Three distinct areas of tissue and at least 1500 cells were analysed per section. Data presented are the % cells showing moderate+strong RelAp536 staining, as indicated by image analysis software. * Significant (P < 0.05) difference between the % stained cells in treated and non-treated sections. (F) Graph showing the relationship between aspirin-induced changes in TIF-IA and RelAp536 staining for six individual tumours. Pearsons correlation coefficient (r2) was used to examine the relationship. (G) Proposed model. Generation of ceramide by specific stresses inhibits CDK4 kinase activity, which induces degradation of TIF-IA in a manner dependent on UBF/p14ARF. The consequent disruption of the PolI pre-initiation complex causes distinct changes in nucleolar architecture and triggers a protein or pathway (as yet unknown) which causes phosphorylation and degradation of IκBα, nuclear translocation of RelA and ultimately, transcription of a gene programme that alters cell phenotype. We propose the nature of this transcriptional program is cell type and stimulus dependent.