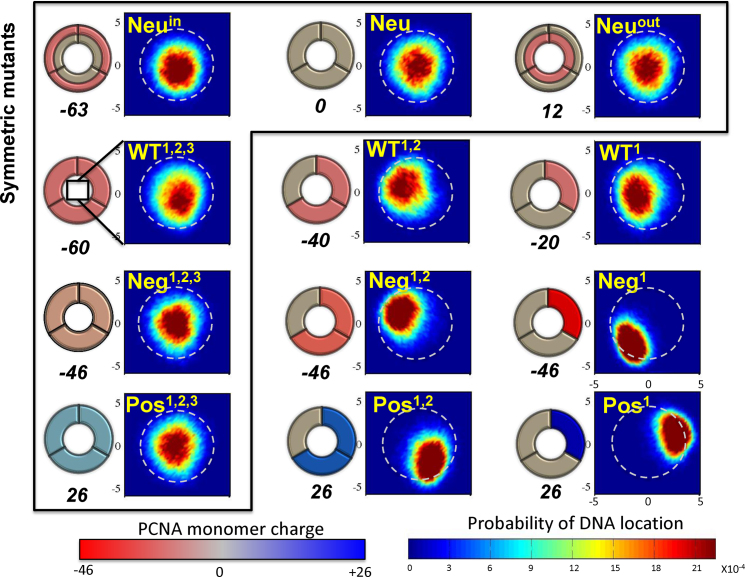
Figure 5.
Schematics and localization probability maps for the DNA center of mass inside the inner rings of the PCNA variants. The Neu set includes variants in which different regions were neutralized by reparametrizing the charges of charged amino acids: the whole protein (Neu), the inner ring (Neuin), or the outer ring (Neuout). The differences within each of the other three sets (WT, Neg, Pos) were achieved by manipulating zero, one, or two subunits of the trimeric PCNA. The WT set includes variants whose monomers bear the same charges as the WT PCNA on three, two, or one subunit(s). The Neg and Pos sets include variants in which the positive or negative charges were neutralized, respectively. Thus, the charge on charged residues becomes more negative (decreasing from –0.33 to –1.0) across the Neg row and similarly becomes increasingly positive (from +0.33 to +1.0) across the Pos row. This is indicated by the increasingly intense shade of the charged subunit in the schematic drawings of the PCNA rings. The colors of the subunits in the schematics are based on the monomer net charge (red to blue color bar) and the overall net charge of each variant appears under its diagram. The variants inside the frame are symmetric whereas those outside the frame are asymmetric. The WT1,2,3 variant corresponds to wild-type PCNA. A probability map for DNA localization within the PCNA ring is shown to the right of each schematic, with probability indicated by the rainbow color bar. A direct interaction between the DNA and PCNA is expected to occur at R ∼ 4 Å (deduced from the interaction of variant Pos1 with DNA), as indicated by the dashed circular line on each probability map.