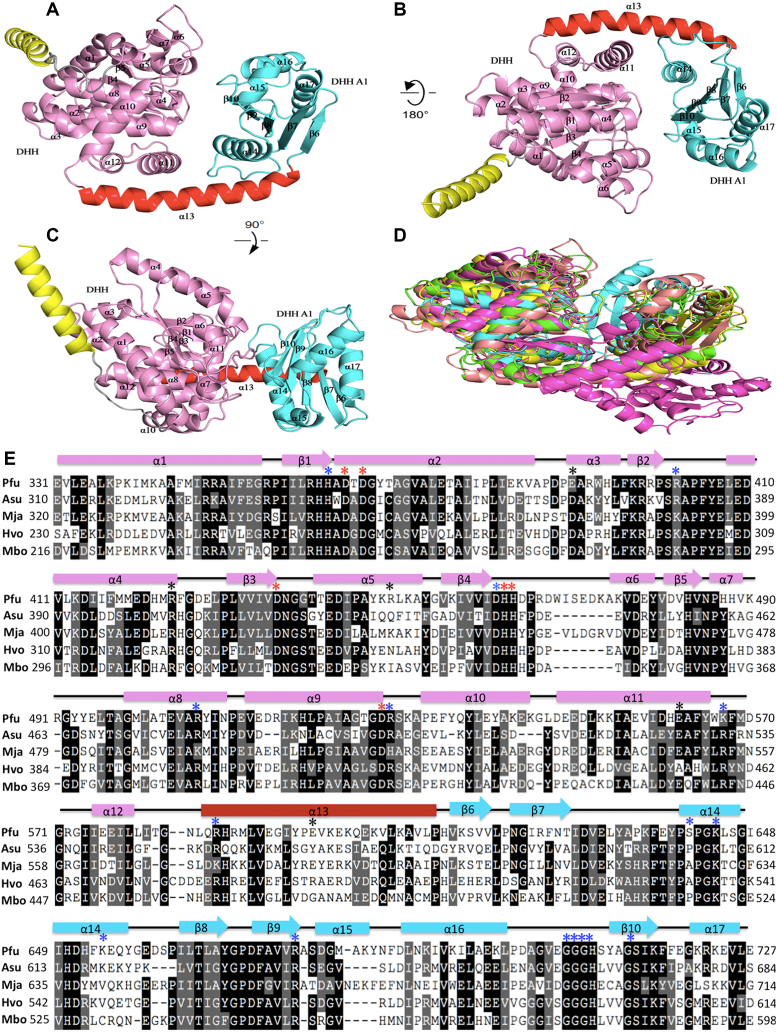
Figure 4.
Crystal structures of PfuHAN_DND and other DHH phosphoesterases. (A) The overall structure of PfuHAN_DND is shown by rotation of (B) 180° and (C) 90°. The DHH and DHHA1 domains are shown in pink and cyan, respectively. The long alpha helix connecting DHH and DHHA1 domains is shown in red. The first alpha helix belongs to the OB-fold domain and is shown in yellow. (D) Comparison of the structures of DHH superfamily members. The structures of PfuHAN_DND (pink), catalytic core (DHH and DHHA1 domain) of Thermus thermophiles RecJ (green, PDB 1IR6), TkoGAN (purple, PDB 5GHT), Streptococcus gordonii PPase (cyan, PDB 1K20), and BsuNrnA (yellow, PDB 5J21) are superposed. (E) Multiple sequence alignment of the HAN DHH-DHHA1 nuclease domains. The alignment was performed using multiple sequence alignment programmes ClustalW and was manually modified to take into account the secondary structural prediction. Completely and partially conserved residues are shaded in dark and light, respectively. Secondary structural elements in PfuHAN_DND are represented at the top of the sequences. The horizontal cylinders indicate α-helices, and the arrows indicate β-strands. The mutated conserved residues for coordinating metal ions (D364, D366, D436, H462, H463 and D528), binding substrates (H362, R402, D461, R507, R529, K567, R587, S641, K644, K654, R676, G706, G707, G708, H709 and S714), and forming trimeric complexes (E390, R449, E598, R424, and E562) are indicated with red, blue, and black asterisks, respectively, above the PfuHAN sequence. The HANs are from Pyrococcus furiosus (Pfu, PF0399), Methanocaldococcus jannaschii (Mja, MJ1198), Haloferax volcanii (Hvo, Hvo1018), Archaeoglobus sulfaticallidus (Asu, Asulf01445) and Methanoregula boonei (Mbo, Mboo1586).