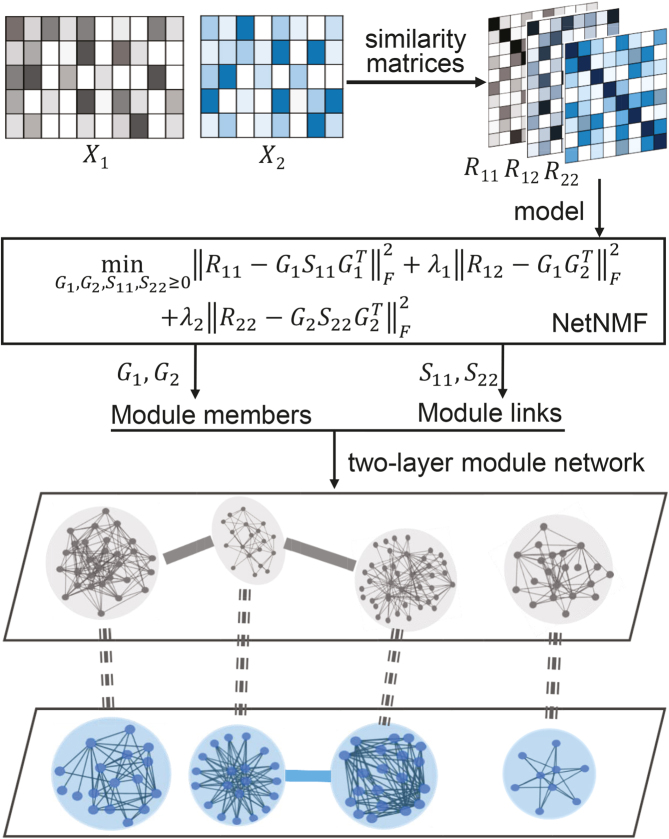
Figure 1.
Overview of the NetNMF for discovering a two-level module network by integrating pairwise genomic data. Three matrices R11, R12, R22 are computed via Pearson correlation, representing the similarities within and between two types of features in the pairwise input data matrices X1 and X2. NetNMF simultaneously decomposes R11, R12, R22 to get the underlying co-modules and their associations. The ith co-module is identified based on the ith column vector in factored matrices G1 and G2; the association degree between the ith and jth modules is determined by S11(i, j) (or S22(i, j)), where S(i, j) represents the element of the ith row and jth column in this matrix. Thus, a two-layer module network could be constructed in which a node represents a module.