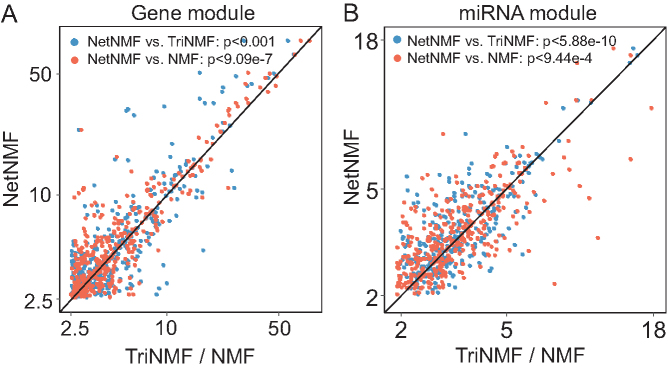
Figure 4.
Comparison of all the enriched GO BPs of modules detected by NetNMF, TriNMF and NMF using TCGA breast cancer dataset. NetNMF performs better than TriNMF and NMF via one-sided Wilcoxon signed rank tests. (A) Comparison for gene modules. For each GO BP, we compute enrichment scores (−log10(P-value)) and the highest score among all modules is taken as the final score of this GO BP for each method. The scores for NetNMF are plotted against those of TriNMF and NMF. Majority of terms are above the central diagonal line, that is, they are more significantly enriched using NetNMF than TriNMF (59%) or NMF (66%). (B) Comparison for the target gene set of miRNA modules. Similar setting with (A). About 60 and 57% of terms are above the central diagonal line comparing NetNMF with TriNMF and NMF, respectively.