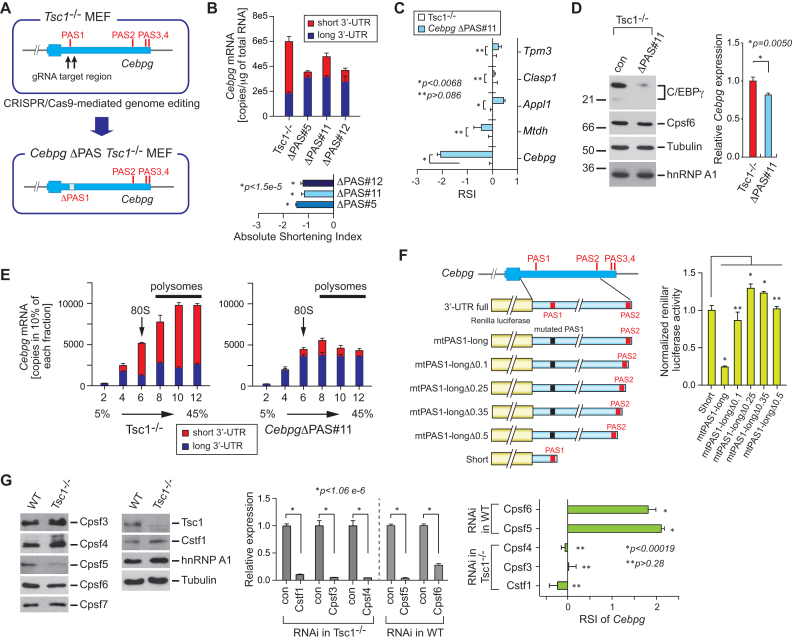
Figure 5.
A regulatory role of 3′-UTR APA in Cebpg gene expression. (A) Schematic of proximal PAS knockout (KO) in Tsc1−/− MEFs. Using the CRISPR/Cas9-mediated genome editing tool, two juxtaposed proximal PASs in the 3′-UTR of Cebpg were deleted and CebpgΔPAS Tsc1−/− MEFs were established. (B) An absolute quantitation of total Cebpg mRNAs and Cebpg mRNA with long 3′-UTR was measured and presented by copies/μg total RNAs. The quantitation was made using three CebpgΔPAS Tsc1−/− MEF clones (#5, #11 and #12). The shortening index of Cebpg in these clones was calculated based on the absolute quantitation. qPCR results are from four technical replicates. Data represent the mean ± SEM. Two-tailed Student's t-tests were performed for statistical significance. (C) The RSI of selected genes in CebpgΔPAS#11 Tsc1−/− MEFs compared to control Tsc1−/− MEFs was measured using qPCR. qPCR results are from four technical replicates. Data represent the mean ± SEM. Two-tailed Student's t-tests were performed for statistical significance. (D) CebpgΔPAS Tsc1−/− MEFs were characterized using western blotting. Expression of C/EBPγ, Cpsf6, Tubulin and hnRNP A1 protein was analyzed using total cell extracts from CebpgΔPAS and control Tsc1−/− MEFs. qPCR using the same set of cell lines was performed to measure the relative level of Cebpg mRNA. qPCR results are from 4 technical replicates. Data represent the mean ± SEM. Two-tailed Student's t-tests were performed for statistical significance. (E) Total and long 3′-UTR Cebpg mRNAs were analyzed using polysome profiling. Cytoplasmic extracts from Tsc1−/− and CebpgΔPAS Tsc1−/− MEFs were fractionated by 5–45% sucrose gradients. Percentage of total and long 3′-UTR Cebpg mRNAs was calculated in each fraction using qPCR. 80S monosome and polysomes in the fractionation were indicated. qPCR results are from four technical replicates. Data represent the mean ± SEM. (F) A serial deletion of the Cebpg 3′-UTR was tested for the regulation of Renilla luciferase expression. 100 (Δ0.1), 250 (Δ0.25), 350 (Δ0.35) and 500 (Δ0.5) nucleotide deletion constructs were tested. Renilla luciferase activity was normalized to firefly luciferase activity which was used as a transfection control. The assay was conducted four technical replicates. Three independent experiments showed similar results. One representative result is shown. Data represent the mean ± SEM. Two-tailed Student's t-tests were performed for statistical significance. P-values were obtained by comparing Short versus each deletion construct. *P < 0.00054. **P > 0.05. (G) Western blot analysis of multiple polyadenylation factors using WT and Tsc1−/− MEFs extracts is shown (left panel). RNAi-mediated knockdown of polyadenylation factors in WT MEFs were conducted (middle panel). The knockdown effect of those polyadenylation factors on the RSI was measured by qRT-PCR (right panel).