Figure 2.
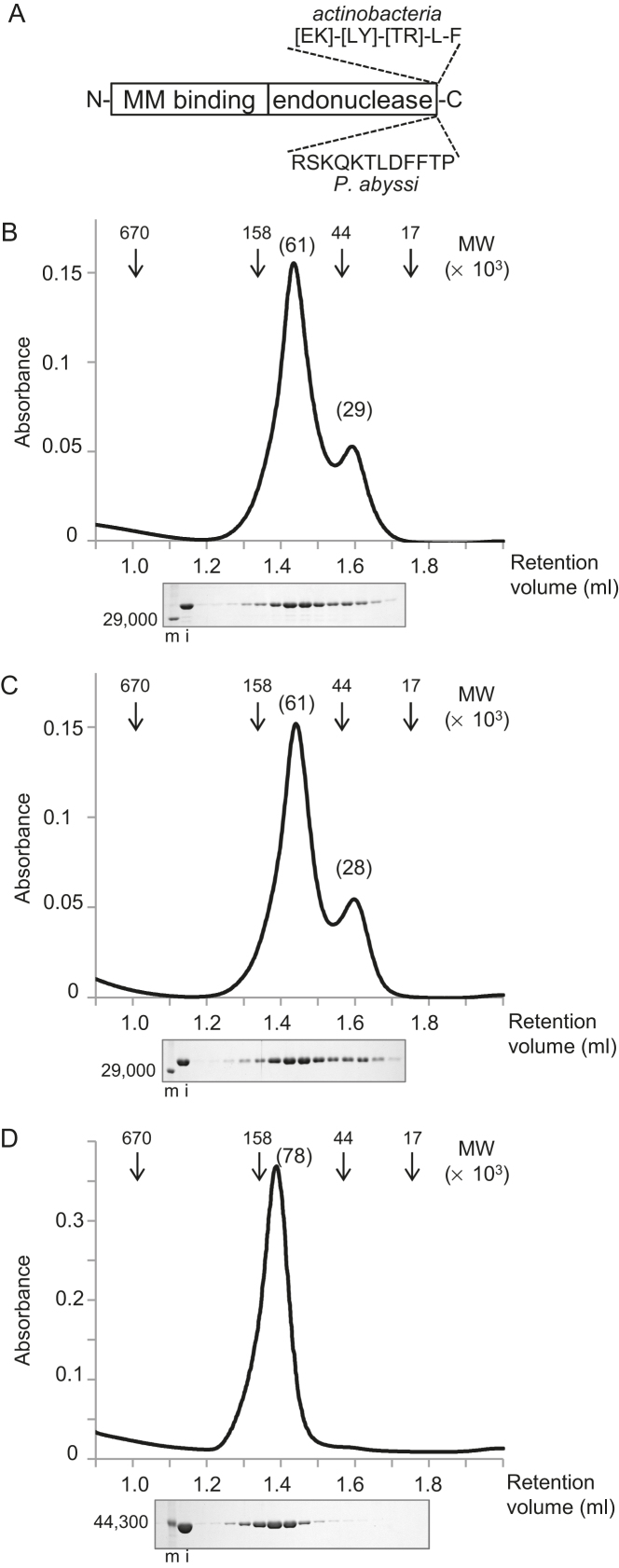
Properties of CglEndoMS-WT and Cglβ-clamp. (A) The two-domain structure of the EndoMS/NucS proteins. ‘MM binding’ and ‘endonuclease’ refer to mismatch binding and endonuclease active site domains, respectively. The positions of the experimentally determined and putative replication clamp binding motif for the Thermococcales and actinobacterial EndoMS/NucS proteins are shown. (B–D) Gel filtration chromatography analyses of CglEndoMS-WT (B), CglEndoMS-ΔCter (C), and Cglβ-clamp (D). Elution profiles, monitored by the absorbance at 280 nm, are shown in the upper part. The arrowheads indicate the elution positions of the standard marker proteins, and the numbers in parentheses on the peak tops indicate the relative molecular masses estimated by the standard curve from the marker proteins. Aliquots of each fraction were subjected to an SDS-12% PAGE followed by Coomassie Brilliant Blue staining (lower part). Protein size markers were run in lanes m, and their sizes are indicated on the left sides of the gels. The loading samples (20%) were run in lanes i.