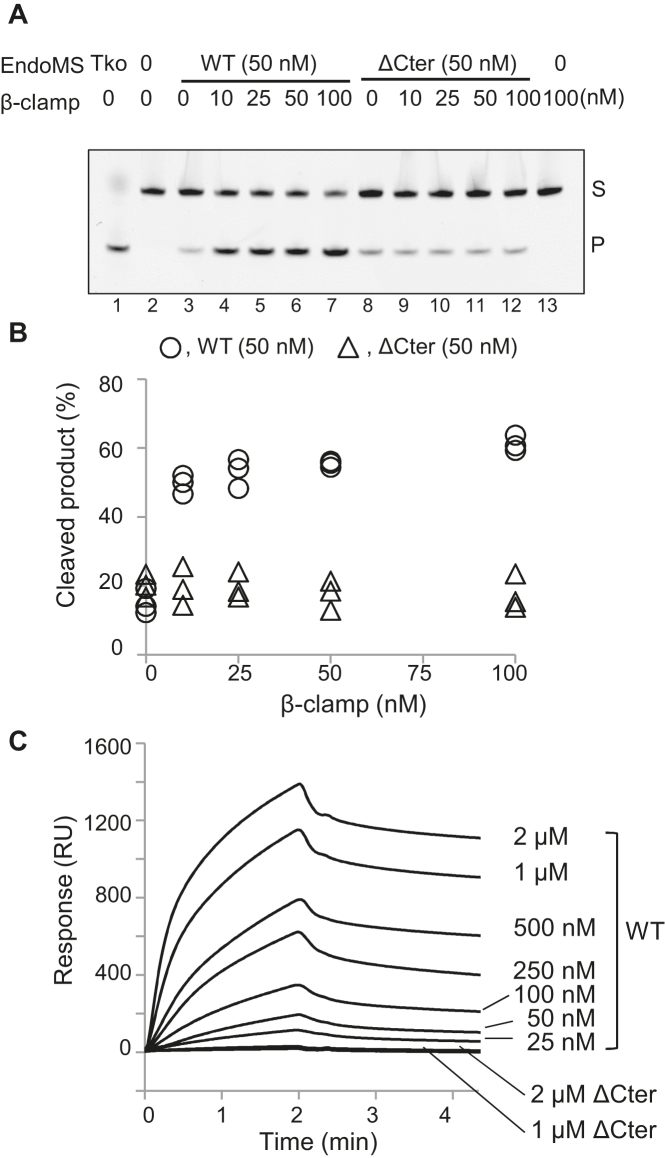
Figure 3.
Cleavage of mismatch-containing DNA by CglEndoMS-WT and CglEndoMS-ΔCter with Cglβ-clamp. (A) The 5′-Cy5-labeled DNA substrates (5 nM) containing the GT base pair were incubated with the proteins. The products were analyzed by native 10% PAGE followed by laser scanning (cropped gel image). Lanes 1, positive control (20 nM TkoEndoMS, as a monomer); 2, negative control (no proteins); 3–7, 50 nM CglEndoMS-WT (as a monomer); 8–12, 50 nM CglEndoMS-ΔCter (as a monomer); 13, no EndoMS. The indicated concentrations (as a monomer) of Cglβ-clamp were added to the reactions. Representative results are shown. The band assignments are indicated on the side of the panels: s, substrates: p, cleaved products. (B) Quantification of the cleaved products. Independent data points from three measurements are plotted. (C) The physical interactions of CglEndoMS with Cglβ-clamp were characterized by SPR using a BIACORE J system (Biacore Inc.). Purified Cglβ-Clamp was immobilized on the sensor Chip CM5, and various concentrations (indicated on the right sides of the sensorgrams) of CglEndoMS-WT and CglEndoMS-ΔCter were loaded onto the chip for 2 min at a flow rate of 30 μl/min in running buffer (10 mM HEPES, pH 7.4, 150 mM NaCl, 3 mM EDTA, and 0.05% surfactant P20). Regeneration was achieved using a 1-min injection of 0.025% SDS. Each background response was subtracted. Data were analyzed using BIAevaluation v.3 software (Biacore Inc.)