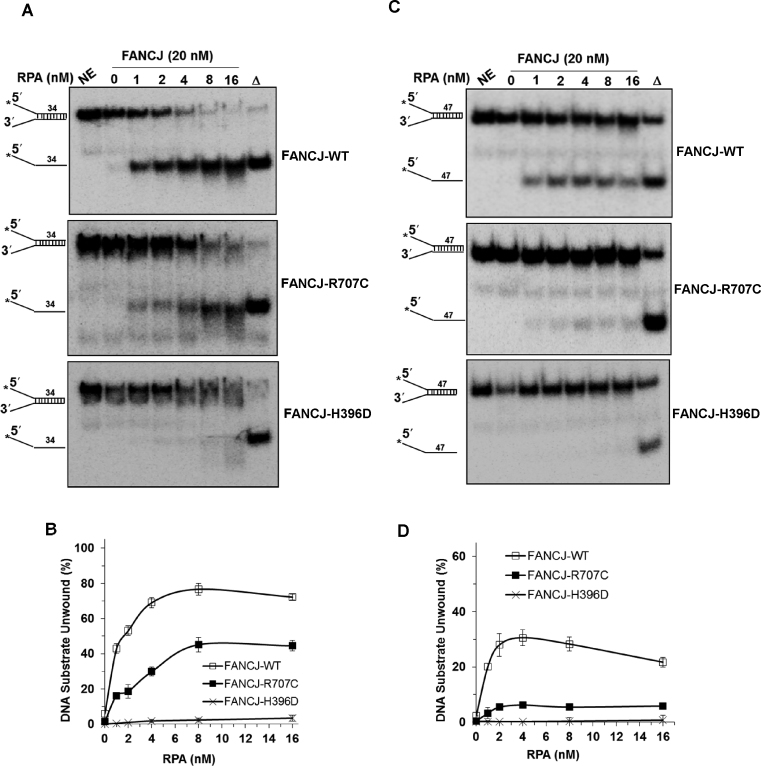
Figure 3.
FANCJ-R707C displays poor unwinding of longer DNA duplexes even in the presence of its interacting partner RPA. Representative native polyacrylamide gel images showing helicase reaction products from the reactions mixtures containing indicated FANCJ proteins (20 nM) in the presence or absence of the specified concentrations of the single-stranded DNA binding protein RPA with 0.5 nM forked duplex DNA substrate (34 bp (A) or 47 bp (C)) incubated at 30°C for 15 min. (B, D) Quantitative analyses of fork duplex unwinding by FANCJ-proteins is shown for 34 bp (B) or 47 bp (D) substrates with S.D. indicated by error bars. Open square, FANCJ-WT; filled square, FANCJ-R707C; open cross, FANCJ-H396D.