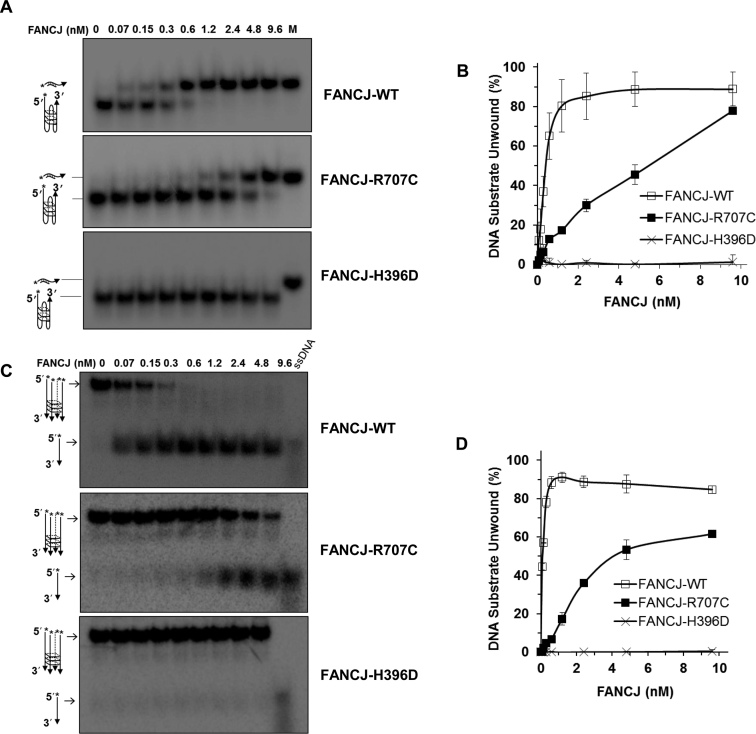
Figure 4.
FANCJ G-quadruplex DNA unwinding is abolished by the H396D mutation but only partially compromised by the R707C mutation. (A, B) Helicase reactions were performed with the indicated FANCJ proteins in the presence of 0.5 nM of Poly (A) Zic1 intramolecular G4 DNA substrate and 5-fold molar excess of complementary PNA oligonucleotide at 30°C for 15 min. Representative gel images of FANCJ reaction mixture products analyzed by native polyacrylamide gel electrophoresis are shown with the conversion of unwound G4 substrate into duplex DNA in Panel A. M represents a positive control marker of duplex DNA generated after annealing of single strand G4 DNA and complementary oligonucleotide. Quantitative analysis of intramolecular G4 unwinding by FANCJ-proteins is shown with S.D, indicated by error bars in Panel B. Open square, FANCJ-WT; filled square, FANCJ-R707C; open cross, FANCJ-H396D. (C, D) Helicase reactions were performed with the indicated FANCJ proteins in the presence of 0.5 nM of tetra-stranded G4 DNA (TPG4) substrate at 30°C for 15 min. Representative gel images of FANCJ reaction mixture products analyzed by native polyacrylamide gel electrophoresis are shown with the conversion of unwound G4 substrate into single-stranded DNA in Panel C. 5′radiolabeled ssDNA as a marker is shown. Quantitative analysis of fork duplex unwinding of FANCJ-proteins is shown with S.D, indicated by error bars in Panel D. Open square, FANCJ-WT; filled square, FANCJ-R707C; open cross, FANCJ-H396D.