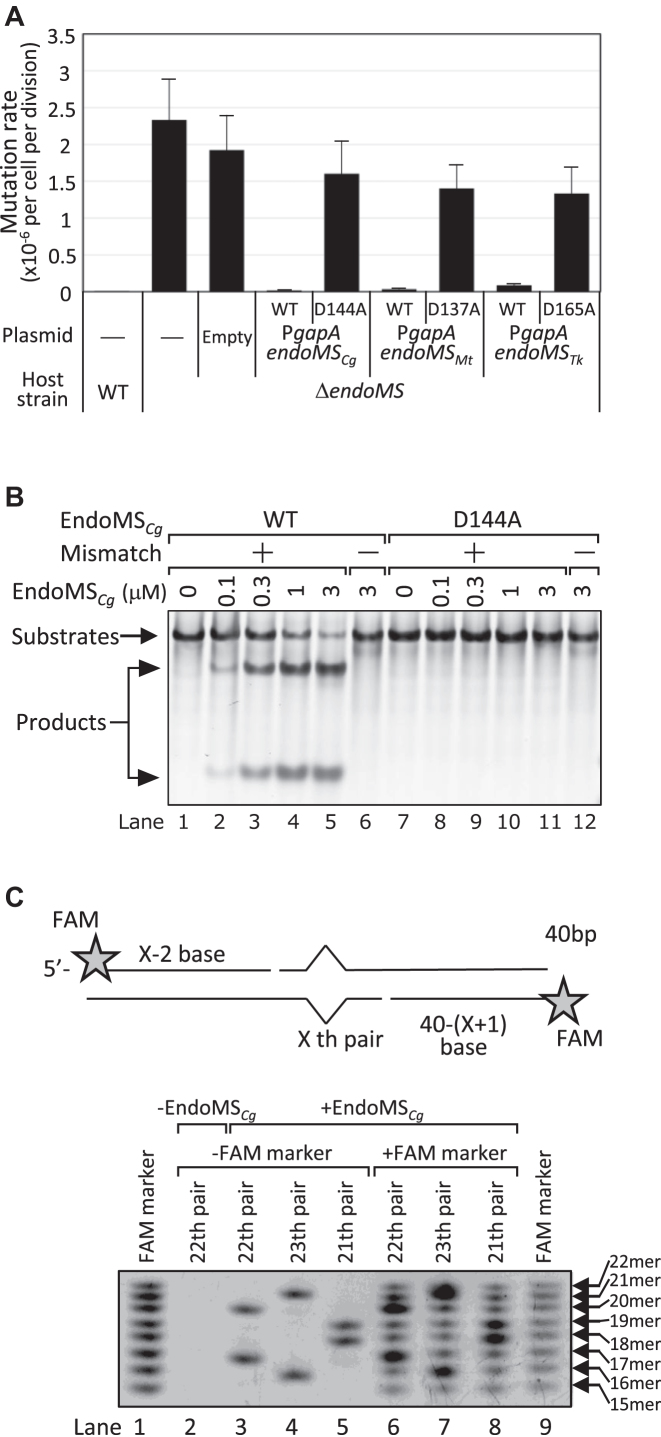
Figure 1.
Complementation of ΔendoMSCg by various EndoMS/NucS overexpression plasmids and in vitro DNA cleavage assay by EndoMS/NucSCg. (A) Rates of spontaneous rifampicin resistance mutations determined by fluctuation assay. Rates of mutations conferring rifampicin resistance to C. glutamicum ATCC13032 (WT) and its ΔendoMSCgderivatives harboring pYTKA1 plasmid with the indicated gene. Error bars show 95% confidence intervals (n = 20). (B, C) In vitro cleavage assay of mismatch-containing DNA by EndoMSCg. (B) DNA substrates with (+) or without (−) G/T mismatch (100-bp, 50 nM) were incubated with the indicated concentrations of wild-type EndoMSCg protein (lane 1–6) or D144A mutant (lane 7–12). (C) The cleavage site was determined using a G/T mismatch substrate (40-bp, 50 nM) with a FAM fluorescent label at the 5′ ends. Schematic drawing of substrates and cleavage products (upper panel). Results of denaturing PAGE of in vitro cleavage products (lower panel). FAM-labeled DNA substrates containing G/T mismatch at indicated position were subjected to in vitro cleavage assay and the products were separated by denaturing PAGE. To determine the cleavage product size, 5′-FAM-labeled oligonucleotides (15–22 mer) were mixed with cleavage products and analyzed (lane 6–8).