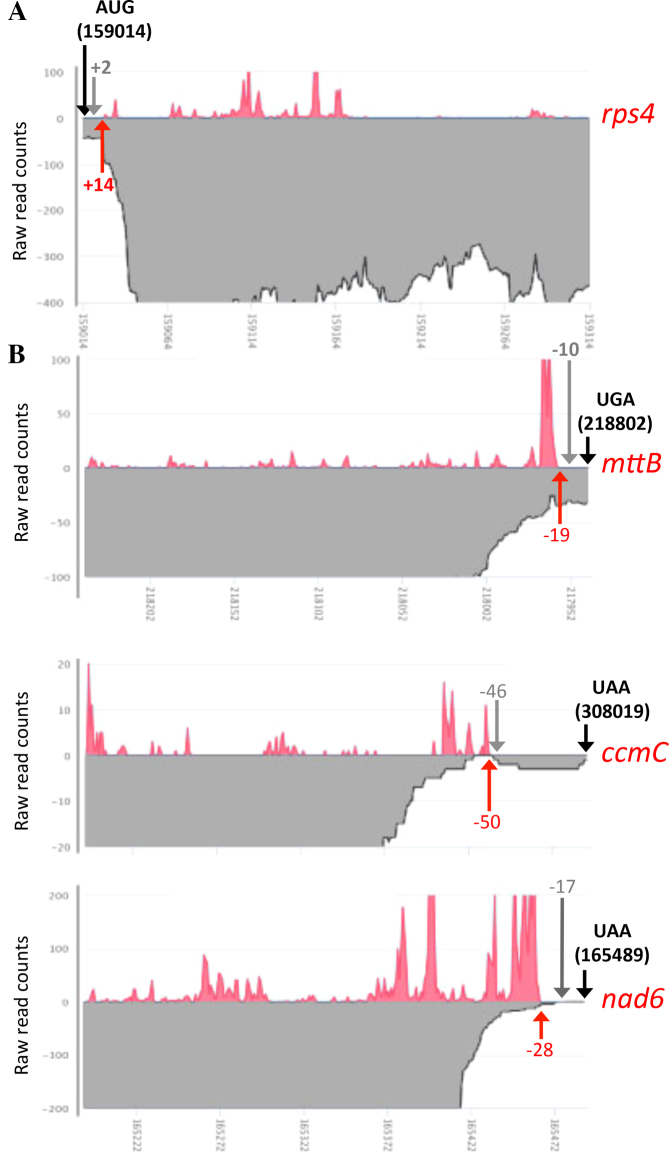
Figure 2.
Ribosome footprint densities near the extremities of Arabidopsis mitochondrial mRNAs for which the start or the stop codons are removed by post-transcriptional processing. Ribosome footprint distributions are shown as red curves and RNA-seq read densities are depicted in gray. (A) Close-up view showing the first 300 nt of the rps4 mRNA. (B) Close-up view showing the last 300 nt of the nad6, mttB and ccmC mRNAs. Black arrows materialize the positions of the theoretical translation start or stop codons and their coordinate on the mitochondrial genome of Col-0 is specified. Red arrows and the associated numbers indicate the start (rps4) or the end (nad6, mttB and ccmC) of observed Ribo-seq signals, respectively to either the translational start or stop codon of each transcript. Gray arrows and the associated numbers indicate the position of the 5′ (rps4) or 3′ (nad6, mttB and ccmC) mRNA extremities mapped by circular RT-PCR in (36,37).