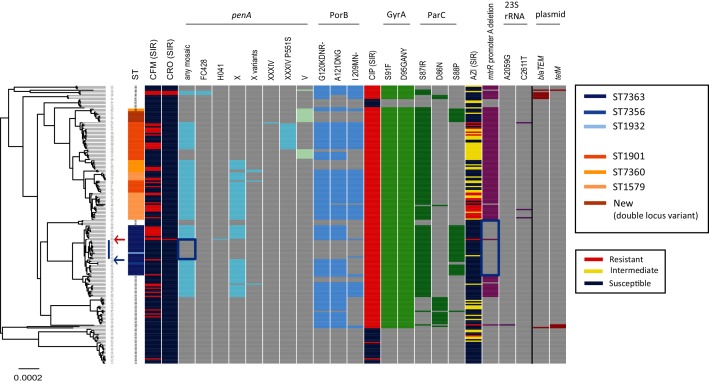
Fig. 1.
Whole-genome sequence phylogeny, resistance patterns of the antimicrobials and genetic polymorphisms. Left: a clonal phylogeny with corrected branch lengths to account for homologous recombination. The blue vertical line represents the notable sub-lineage in ST7363, whereas the blue arrow represents the outgroup of the sub-lineage. The red arrow indicates strain H041 (WHO X). Heatmap: column 1, ST: the two major STs (ST7363 and ST1901), single-locus variants of the former (ST7356, ST1932) and the latter (ST7360 and ST1579), and a new ST (double-locus variant of ST1901) are coloured. Columns 2 and 3: susceptible/resistant (S/R) categories according to the EUCAST MIC breakpoint of cefixime (CFM) and ceftriaxone (CRO). The presence or absence of genetic features is shown in columns 4–14, 16–20, 22–26 as marked: grey indicates absence, other colours indicate presence. Column 4: any mosaic penA allele. Columns 5–10: a specific mosaic penA allele. Column 11: penA V. Columns 12–14: nonsynonymous amino acid changes from wild-types in PorB. Column 15: susceptible/intermediate/resistant (S/I/R) categories according to the EUCAST MIC breakpoints of ciprofloxacin (CIP). Columns 16–17: nonsynonymous amino acid changes from wild-type in GyrA. Columns 18–20: nonsynonymous amino acid changes from wild-type in ParC. Column 21: susceptible/intermediate/resistant (S/I/R) categories according to the EUCAST MIC breakpoints of azithromycin (AZI). Column 22: the adenosine deletion in the mtrR promoter. Columns 23–24: the mutations in the 23S rRNA gene. Columns 25–26: plasmid-encoded blaTEM and tetM. The blue rectangles indicate notable sub-lineages in ST7363 described in the text.