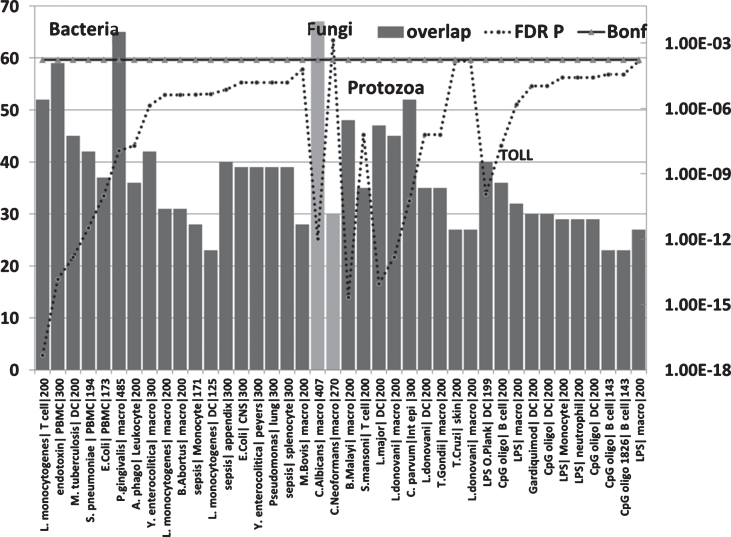
Fig.6.
Overlaps between genes upregulated in the AD hippocampal transcriptome and those upregulated by bacteria, fungi, nematode (B. malayi)/trematode (S. mansoni), protozoan, LPS and TLR ligands in immunocompetent cells (MSigDB or GEO data). (X axis = pathogen/ligand| celltype|N upregulated genes from MSigDb or Geo): Y1 = N overlap; Y2 = p value). Pathogen or ligand abbreviations (from left): L. monocytogenes, Listeria monocytogenes; endotoxin, gram-negative bacterial wall component; S. pneumoniae, Streptococcus pneumoniae; E. coli, Escherichia coli; P. gingivalis, Porphyromonas gingivalis; A. phago, Anaplasma phagocytophilum; Y. enterocolitica, Yersinia enterocolitica; M. bovis, Mycobacterium bovis; C. albicans, Candida albicans; C. neoformans, Cryptococcus neoformans; B. malayi, Brugia malayi (filarial parasite causing elephantiasis); S. mansoni, Schistosoma mansoni; L. donovani, Leishmania donovani; C. parvum, Cryptosporidium parvum; L. major, Leishmania major; T. gondii, Toxoplasma gondii; T. cruzi, Trypanosoma cruzi; LPS, lipopolysaccharide; LPS O.Plank, Oscillatoria Planktothrix (cyanobacterial lipopolysaccharide); CpG oligo, CpG Oligodeoxynucleotide (TLR9 ligand); Gardiquimod, TLR7 ligand. Cell type abbreviations as for Fig. 5: + CNS, central nervous system; peyers, peyers patch; Int epi, intestinal epithelial cells.