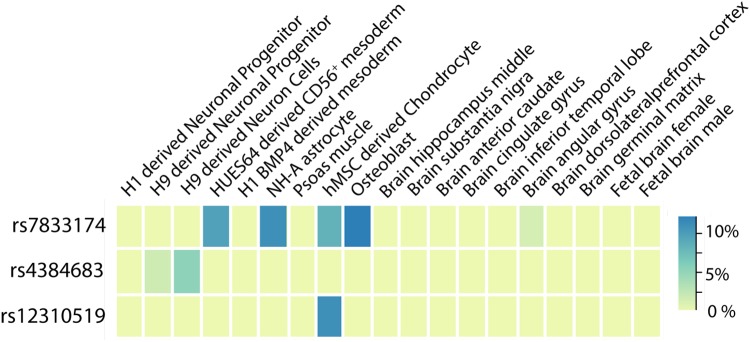
Fig 2. Significant variants are co-localized with potential gene regulatory markers.
The heatmap depicts the percentage of variants in gene regulatory regions (associated with enhancers/promotors) in LD (r2>0.6) with rs7833174 (CCDC26/GSDMC), rs12310519 (SOX5), and rs4384683 (DCC). We examined epigenetic histone marks in selected cell types/tissues including chondrogenic, bone-related, neuronal and brain cells/tissues; mesodermal cells (related to notochord); and psoas muscle (located proximal to the lumbar spine).