Fig. 2.
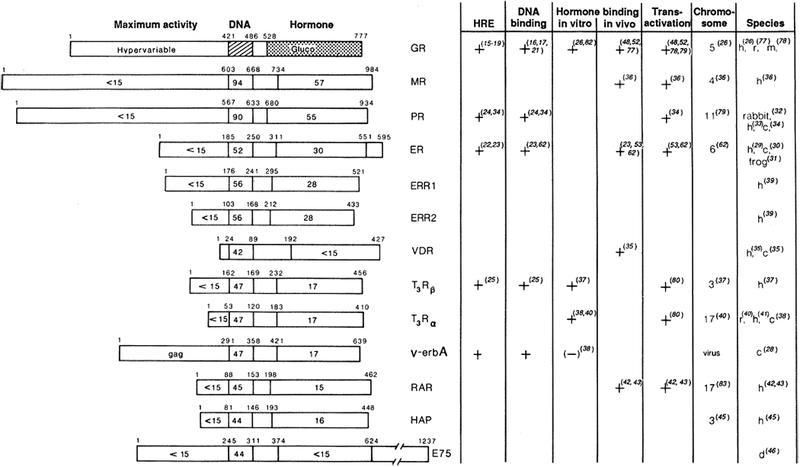
Schematic amino acid comparison of members of the steroid hormone receptor superfamily. Primary amino acid sequences have been aligned on the basis of regions of maximum amino acid similarity, with the percentage amino acid identity indicated for each region in relation to the hGR (55). Domains shown are a domain at the NH2-terminal end, required for “Maximum activity”; the 66- to 68-amino acid DNA-binding core (“DNA”); and the 25-amino acid hormone-binding domain (“Hormone”). The amino acid position of each domain boundary is shown. Amino acid numbers for all receptors represent the human forms with the exception of v-erbA and E75 (46). Functional assignments have been determined by characterization of the glucocorticoid and estrogen receptors. Designations are as follows: GR, glucocorticoid receptor; MR, mineralocorticoid receptor; PR, progesterone receptor; ER, estrogen receptor; ERR1 or ERR2, estrogenr eceptor–related1 or 2; VDR, vitaminD3 receptor;and T3Rβ and T3Rα, thyroid hormone receptors. The (+) or (−) indicates whether a particular property has been demonstrated for the products of cloned receptor cDNA or with purified receptor. HRE, hormone response element. This relates to whether the binding site has been identified structurally and whether its enhancement properties have been demonstrated by gene transfer studies. For PR, DNA-binding properties have been shown only with the native purified receptor. “Hormone binding in vitro” indicates whether this property has been demonstrated by translation in a rabbit reticulocyte lysate system (26). “Hormone binding in vivo” refers to expression of the cloned receptor in transfected cells. “Chromosome” indicates the human chromosome location. Species are as follows: h, human; r, rat; m, mouse; c, chicken; and d, Drosophila.
