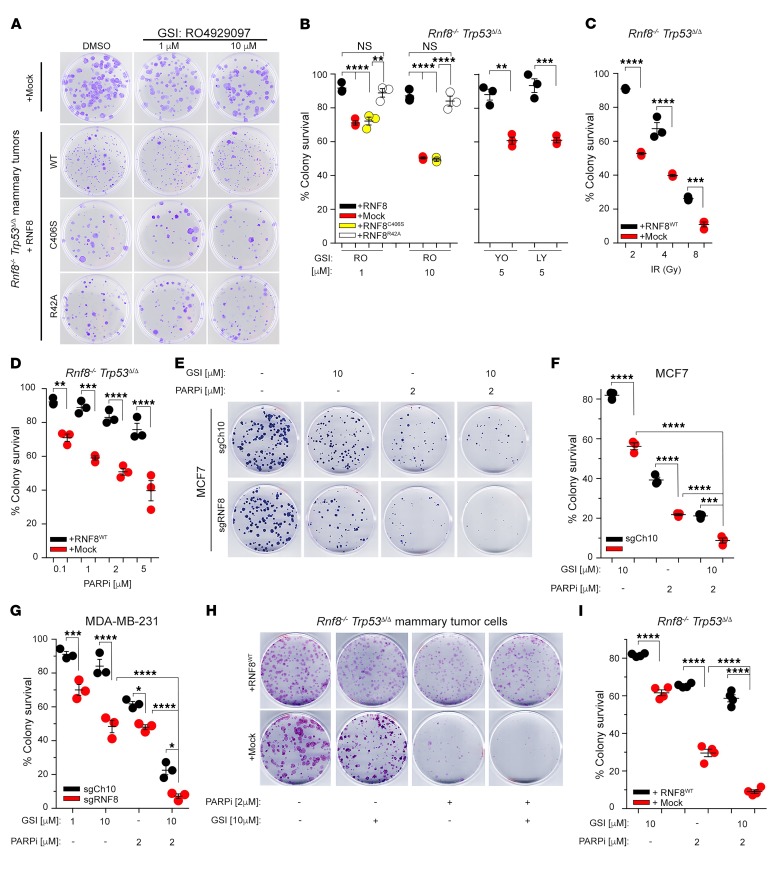
Figure 8. RNF8-deficient breast tumors are sensitive to combination of pharmacological inhibitors of Notch signaling and PARP.
(A) Representative dish images of 3 independent experiments in triplicate showing clonogenic survival under GSI (RO4929097) treatments of Rnf8–/– Trp53Δ/Δ mammary tumor cells reconstituted with mock or either WT or mutant forms of RNF8 as indicated. Cells were fixed and stained with crystal violet at day 12 of culture. (B) Dot plots depicting percentages of clonogenic survival of Rnf8–/– Trp53Δ/Δ mammary tumor cells and their reconstituted controls (as indicated) after 12 days of treatment with different GSIs (RO, RO4929097; YO, YO-01027; LY, LY411575). (C and D) Dot plots depicting clonogenic survival of Rnf8–/– Trp53Δ/Δ mammary tumor cells, reconstituted with mock or RNF8, 12 days after IR (C) or treatment with the PARPi KU0058948 (D). (E) Representative dish images showing clonogenic survival of MCF7 cells deficient for RNF8 (sgRNF8) and their controls (sgCh10) 25 days after treatment with GSI (RO4929097), PARPi (olaparib), or their combination as indicated. Images are representative of 3 independent experiments in triplicate. (F and G) Dot plots depicting percentages of clonogenic survival of MCF7 cells (F) and MDA-MB-231 cells (G) deficient for RNF8 (sgRNF8) and their controls (sgCh10) after treatment with GSI (RO4929097), PARPi (olaparib), or their combination as indicated. (H) Representative images of clonogenic survival of the indicated mammary tumor cells 12 days after treatment with GSI (RO4929097), PARPi (olaparib), or their combination as indicated. (I) Dot plots depicting clonogenic survival of Rnf8–/– Trp53Δ/Δ mammary tumor cells reconstituted with mock or RNF8 after treatment (12 days) with GSI (RO4929097), PARPi (olaparib), or their combination as indicated. B–D, F, G, and I: n = 3; mean ± SEM, normalized to DMSO-treated controls except for C, in which normalization was to nonirradiated controls. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, 2-way ANOVA followed by Tukey’s test.