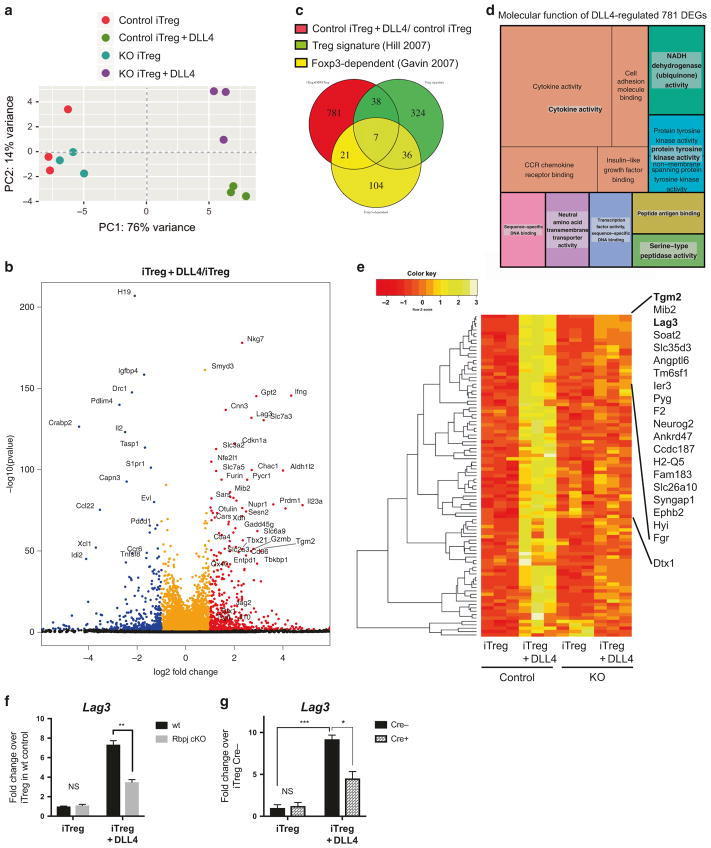
Fig. 7.
Smyd3 deletion changed gene expression profile in the presence of DLL4 stimulation during iTreg differentiation in vitro. a Principle component analysis of RNA-seq data sets of iTreg cells from Cre− control or Cre + Smyd3 cKO with or without DLL4 stimulation, assessed using the top 500 genes with the highest variance. Each symbol represented a single mouse. Each group have three biological triplicates. b Volcano plot showed the differentially expressed genes (DEGs) in control iTreg + DLL4 over control iTreg; red dots indicated genes that were significantly (BH-adjusted p-value < 0.05) upregulated by DLL4 more than twofold (log2FoldChange > 1) and blue dots represents genes that were significantly downregulated by DLL4 more than twofold. The y axis represented the significance of the differential analysis. c Venn diagram demonstrated the overlap between current DLL4-regulated DEGs and published Treg signatures and Foxp3-dependent genes. d Gene Ontology (GO) of un-overlapped DLL4-regulated DEGs (781 genes). Treemap showed the significant GO term with False-discovery-rate (FDR)-adjusted global P-value < 0.05. P-value inversely proportional to area for each GO term. e Normalized expression of all the DEGs that were significantly upregulated by DLL4 stimulation for more than 2.00-fold, and downregulated more than 1.51-fold by Smyd3 deletion. Heatmap presented Z-score in a dendrogram clustered by Pearson’s distribution and ranked based on rowMeans. Top 20 DEGs and Deltex1 were indicated. f Naive CD4 T cells from wild-type control or Rbpj cKO were differentiated to iTreg with or without DLL4 for 24 h. Lag3 expression were detected. g Naive CD4 T cells from Cre− control or Smyd3 cKO were differentiated to iTreg with or without DLL4 for 48 h. Lag3 expression were detected